kmeans.split 20 - GCRMA
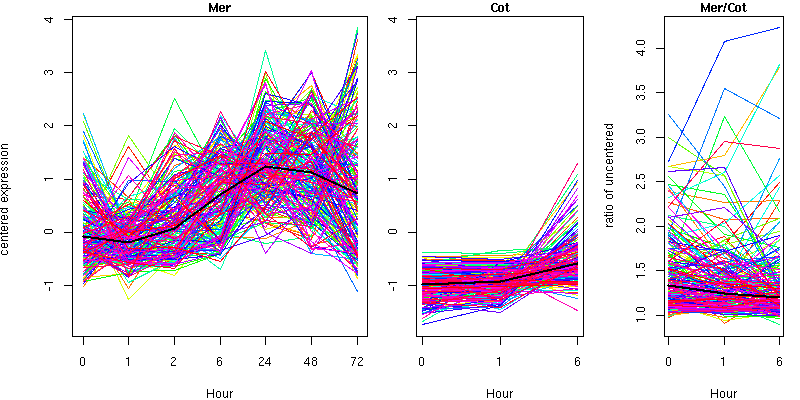
Cluster 1 - contains 208 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005372 | water transporter activity | 4 | 0.28 | 0.00017 |
| GO:0015250 | water channel activity | 4 | 0.28 | 0.00017 |
| GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups | 5 | 1.08 | 0.00467 |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds | 9 | 3.52 | 0.00920 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0042221 | response to chemical stimulus | 28 | 9.39 | 2.2e-07 |
| GO:0009628 | response to abiotic stimulus | 35 | 13.85 | 3.1e-07 |
| GO:0009719 | response to endogenous stimulus | 23 | 7.33 | 1.2e-06 |
| GO:0009733 | response to auxin stimulus | 12 | 2.13 | 1.6e-06 |
| GO:0009725 | response to hormone stimulus | 18 | 5.16 | 4.6e-06 |
| GO:0050896 | response to stimulus | 41 | 20.37 | 9.3e-06 |
| GO:0009605 | response to external stimulus | 11 | 2.54 | 5.2e-05 |
| GO:0007242 | intracellular signaling cascade | 13 | 3.81 | 1.2e-04 |
| GO:0007154 | cell communication | 20 | 7.88 | 1.3e-04 |
| GO:0008361 | regulation of cell size | 7 | 1.31 | 3.5e-04 |
| GO:0000902 | cellular morphogenesis | 7 | 1.49 | 7.6e-04 |
| GO:0009653 | morphogenesis | 10 | 2.94 | 7.7e-04 |
| GO:0044255 | cellular lipid metabolism | 11 | 3.48 | 7.9e-04 |
| GO:0007264 | small GTPase mediated signal transduction | 5 | 0.78 | 1.1e-03 |
| GO:0007165 | signal transduction | 17 | 7.36 | 1.2e-03 |
| GO:0009826 | unidimensional cell growth | 5 | 0.95 | 2.7e-03 |
| GO:0009987 | cellular process | 113 | 93.68 | 3.0e-03 |
| GO:0040007 | growth | 6 | 1.51 | 4.2e-03 |
| GO:0008610 | lipid biosynthesis | 8 | 2.60 | 4.8e-03 |
| GO:0009753 | response to jasmonic acid stimulus | 5 | 1.12 | 5.4e-03 |
| GO:0006629 | lipid metabolism | 12 | 5.11 | 5.4e-03 |
| GO:0007582 | physiological process | 118 | 100.51 | 6.4e-03 |
| GO:0050875 | cellular physiological process | 108 | 90.53 | 6.7e-03 |
| GO:0009861 | jasmonic acid and ethylene-dependent systemic resistance | 5 | 1.26 | 8.7e-03 |
| GO:0016049 | cell growth | 5 | 1.27 | 9.0e-03 |
|
list of genes (text format)
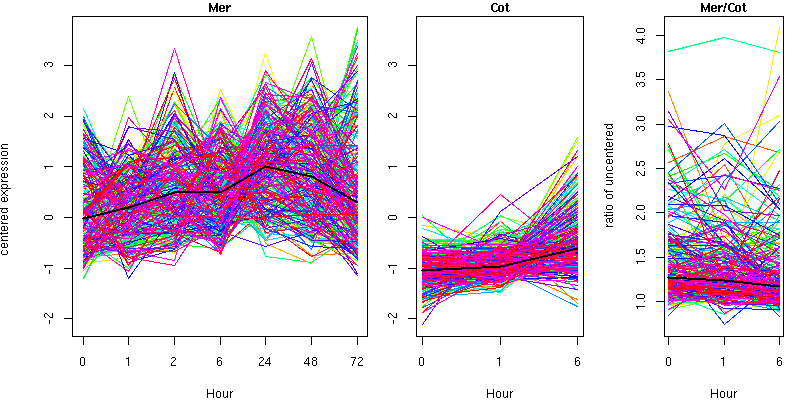
Cluster 2 - contains 271 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016491 | oxidoreductase activity | 26 | 16 | 0.0085 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0007275 | development | 28 | 12.06 | 3.0e-05 |
| GO:0005984 | disaccharide metabolism | 7 | 0.96 | 5.0e-05 |
| GO:0009653 | morphogenesis | 13 | 3.65 | 8.2e-05 |
| GO:0000902 | cellular morphogenesis | 9 | 1.85 | 1.0e-04 |
| GO:0040007 | growth | 9 | 1.88 | 1.1e-04 |
| GO:0016049 | cell growth | 8 | 1.58 | 1.9e-04 |
| GO:0008361 | regulation of cell size | 8 | 1.63 | 2.3e-04 |
| GO:0008202 | steroid metabolism | 5 | 0.57 | 2.6e-04 |
| GO:0009725 | response to hormone stimulus | 17 | 6.41 | 2.6e-04 |
| GO:0016052 | carbohydrate catabolism | 8 | 1.80 | 4.6e-04 |
| GO:0044275 | cellular carbohydrate catabolism | 8 | 1.80 | 4.6e-04 |
| GO:0009828 | cell wall loosening (sensu Magnoliophyta) | 4 | 0.39 | 5.9e-04 |
| GO:0009827 | cell wall modification (sensu Magnoliophyta) | 4 | 0.43 | 8.5e-04 |
| GO:0007582 | physiological process | 149 | 124.71 | 8.6e-04 |
| GO:0006694 | steroid biosynthesis | 4 | 0.44 | 9.6e-04 |
| GO:0050896 | response to stimulus | 41 | 25.28 | 1.2e-03 |
| GO:0006066 | alcohol metabolism | 9 | 2.63 | 1.4e-03 |
| GO:0042221 | response to chemical stimulus | 23 | 11.65 | 1.5e-03 |
| GO:0009414 | response to water deprivation | 6 | 1.25 | 1.6e-03 |
| GO:0009825 | multidimensional cell growth | 4 | 0.51 | 1.6e-03 |
| GO:0046352 | disaccharide catabolism | 4 | 0.51 | 1.6e-03 |
| GO:0046351 | disaccharide biosynthesis | 4 | 0.52 | 1.8e-03 |
| GO:0009987 | cellular process | 139 | 116.25 | 1.8e-03 |
| GO:0006629 | lipid metabolism | 15 | 6.34 | 1.9e-03 |
| GO:0042398 | amino acid derivative biosynthesis | 7 | 1.76 | 2.0e-03 |
| GO:0009719 | response to endogenous stimulus | 19 | 9.09 | 2.0e-03 |
| GO:0009628 | response to abiotic stimulus | 30 | 17.19 | 2.0e-03 |
| GO:0009415 | response to water | 6 | 1.35 | 2.4e-03 |
| GO:0050875 | cellular physiological process | 133 | 112.33 | 4.1e-03 |
| GO:0044255 | cellular lipid metabolism | 11 | 4.32 | 4.3e-03 |
| GO:0009605 | response to external stimulus | 9 | 3.15 | 4.6e-03 |
| GO:0006869 | lipid transport | 5 | 1.09 | 4.8e-03 |
| GO:0009733 | response to auxin stimulus | 8 | 2.64 | 5.2e-03 |
| GO:0008610 | lipid biosynthesis | 9 | 3.23 | 5.3e-03 |
| GO:0009826 | unidimensional cell growth | 5 | 1.19 | 6.7e-03 |
| GO:0009664 | cell wall organization and biogenesis (sensu Magnoliophyta) | 5 | 1.20 | 7.1e-03 |
| GO:0019752 | carboxylic acid metabolism | 14 | 6.68 | 7.5e-03 |
| GO:0042545 | cell wall modification | 5 | 1.22 | 7.7e-03 |
| GO:0006082 | organic acid metabolism | 14 | 6.71 | 7.8e-03 |
| GO:0007047 | cell wall organization and biogenesis | 7 | 2.30 | 8.6e-03 |
| GO:0045229 | external encapsulating structure organization and biogenesis | 7 | 2.30 | 8.6e-03 |
| GO:0006575 | amino acid derivative metabolism | 7 | 2.30 | 8.6e-03 |
|
list of genes (text format)
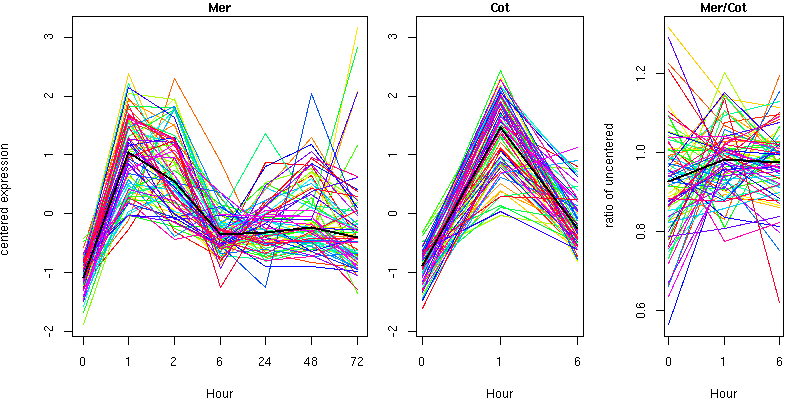
Cluster 3 - contains 71 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009651 | response to salt stress | 5 | 0.52 | 0.00017 |
| GO:0006970 | response to osmotic stress | 5 | 0.58 | 0.00028 |
| GO:0009628 | response to abiotic stimulus | 12 | 4.35 | 0.00108 |
| GO:0009416 | response to light stimulus | 5 | 0.80 | 0.00122 |
| GO:0009314 | response to radiation | 5 | 0.81 | 0.00129 |
| GO:0006950 | response to stress | 10 | 3.55 | 0.00255 |
| GO:0050789 | regulation of biological process | 13 | 5.51 | 0.00269 |
| GO:0050896 | response to stimulus | 14 | 6.40 | 0.00362 |
| GO:0050791 | regulation of physiological process | 12 | 5.14 | 0.00437 |
| GO:0009605 | response to external stimulus | 4 | 0.80 | 0.00831 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
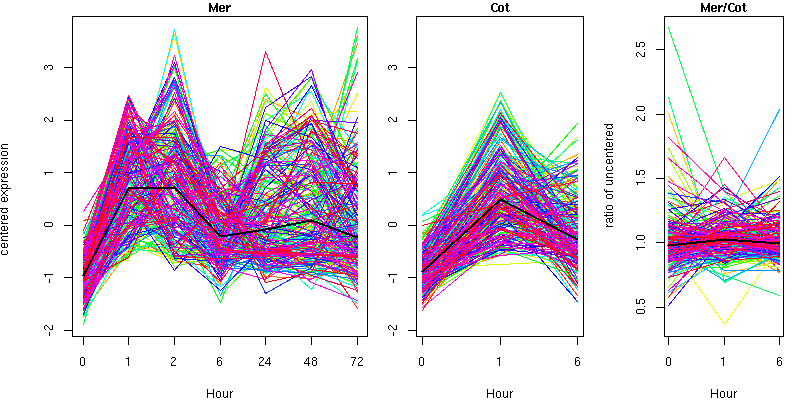
Cluster 4 - contains 218 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003700 | transcription factor activity | 28 | 14.8 | 0.00079 |
| GO:0030528 | transcription regulator activity | 30 | 16.3 | 0.00082 |
| GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups | 5 | 1.1 | 0.00577 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009628 | response to abiotic stimulus | 41 | 13.85 | 2.4e-10 |
| GO:0050896 | response to stimulus | 50 | 20.37 | 1.1e-09 |
| GO:0006950 | response to stress | 31 | 11.30 | 2.9e-07 |
| GO:0042221 | response to chemical stimulus | 27 | 9.39 | 7.5e-07 |
| GO:0009408 | response to heat | 7 | 0.69 | 5.8e-06 |
| GO:0019748 | secondary metabolism | 13 | 3.11 | 1.5e-05 |
| GO:0009814 | defense response to pathogen, incompatible interaction | 10 | 1.92 | 2.4e-05 |
| GO:0009861 | jasmonic acid and ethylene-dependent systemic resistance | 8 | 1.26 | 3.9e-05 |
| GO:0009611 | response to wounding | 9 | 1.75 | 6.9e-05 |
| GO:0009753 | response to jasmonic acid stimulus | 7 | 1.12 | 1.4e-04 |
| GO:0009719 | response to endogenous stimulus | 19 | 7.33 | 1.4e-04 |
| GO:0048511 | rhythmic process | 4 | 0.29 | 2.0e-04 |
| GO:0007623 | circadian rhythm | 4 | 0.29 | 2.0e-04 |
| GO:0051707 | response to other organism | 15 | 5.18 | 2.3e-04 |
| GO:0009605 | response to external stimulus | 10 | 2.54 | 2.5e-04 |
| GO:0009416 | response to light stimulus | 10 | 2.55 | 2.5e-04 |
| GO:0009651 | response to salt stress | 8 | 1.66 | 2.7e-04 |
| GO:0009314 | response to radiation | 10 | 2.58 | 2.8e-04 |
| GO:0006970 | response to osmotic stress | 8 | 1.85 | 5.5e-04 |
| GO:0010038 | response to metal ion | 5 | 0.69 | 6.4e-04 |
| GO:0009613 | response to pest, pathogen or parasite | 13 | 4.52 | 6.5e-04 |
| GO:0009751 | response to salicylic acid stimulus | 6 | 1.05 | 6.5e-04 |
| GO:0042829 | defense response to pathogen | 11 | 3.45 | 7.3e-04 |
| GO:0009607 | response to biotic stimulus | 16 | 6.51 | 8.6e-04 |
| GO:0009411 | response to UV | 4 | 0.45 | 1.1e-03 |
| GO:0010035 | response to inorganic substance | 5 | 0.81 | 1.3e-03 |
| GO:0009733 | response to auxin stimulus | 8 | 2.13 | 1.4e-03 |
| GO:0009723 | response to ethylene stimulus | 6 | 1.22 | 1.4e-03 |
| GO:0046686 | response to cadmium ion | 4 | 0.49 | 1.5e-03 |
| GO:0009266 | response to temperature stimulus | 8 | 2.18 | 1.6e-03 |
| GO:0009739 | response to gibberellic acid stimulus | 5 | 0.91 | 2.2e-03 |
| GO:0042828 | response to pathogen | 11 | 4.16 | 3.2e-03 |
| GO:0009725 | response to hormone stimulus | 12 | 5.16 | 5.9e-03 |
| GO:0006952 | defense response | 12 | 5.16 | 5.9e-03 |
| GO:0006720 | isoprenoid metabolism | 5 | 1.15 | 6.1e-03 |
| GO:0016096 | polyisoprenoid metabolism | 4 | 0.78 | 7.6e-03 |
| GO:0006721 | terpenoid metabolism | 4 | 0.78 | 7.6e-03 |
| GO:0042214 | terpene metabolism | 4 | 0.79 | 8.0e-03 |
|
list of genes (text format)
Cluster 5 - contains 166 genes
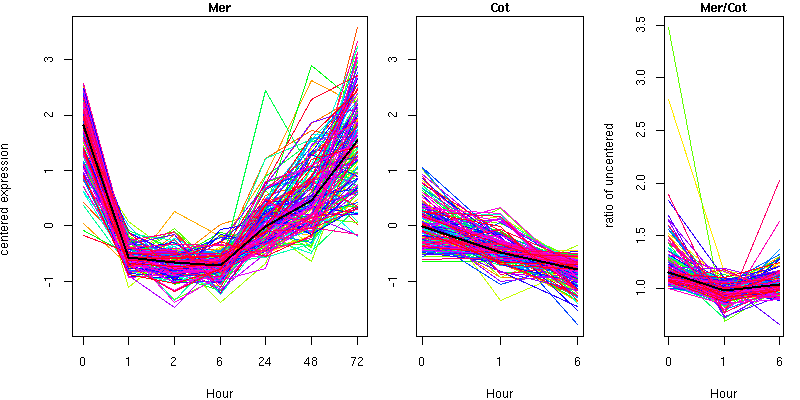
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005478 | intracellular transporter activity | 4 | 0.26 | 0.00012 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006944 | membrane fusion | 5 | 0.25 | 4.6e-06 |
| GO:0016044 | membrane organization and biogenesis | 5 | 0.37 | 3.2e-05 |
| GO:0042157 | lipoprotein metabolism | 8 | 2.82 | 7.5e-03 |
| GO:0042158 | lipoprotein biosynthesis | 8 | 2.82 | 7.5e-03 |
| GO:0006497 | protein amino acid lipidation | 8 | 2.82 | 7.5e-03 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0008372 | cellular component unknown | 67 | 47.36 | 0.00042 |
| GO:0044431 | Golgi apparatus part | 4 | 0.47 | 0.00127 |
| GO:0005794 | Golgi apparatus | 4 | 0.61 | 0.00330 |
|
list of genes (text format)
Cluster 6 - contains 481 genes
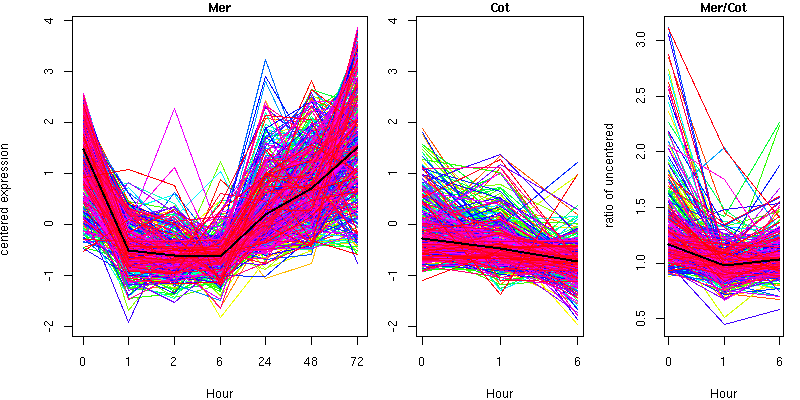
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005515 | protein binding | 66 | 33.26 | 6.2e-08 |
| GO:0005488 | binding | 180 | 144.04 | 1.6e-04 |
| GO:0030528 | transcription regulator activity | 57 | 35.29 | 2.2e-04 |
| GO:0004840 | ubiquitin conjugating enzyme activity | 6 | 0.90 | 2.5e-04 |
| GO:0008270 | zinc ion binding | 35 | 18.94 | 3.7e-04 |
| GO:0008639 | small protein conjugating enzyme activity | 6 | 0.97 | 3.8e-04 |
| GO:0003700 | transcription factor activity | 51 | 31.99 | 6.4e-04 |
| GO:0004707 | MAP kinase activity | 4 | 0.46 | 1.0e-03 |
| GO:0008017 | microtubule binding | 4 | 0.51 | 1.5e-03 |
| GO:0004702 | receptor signaling protein serine/threonine kinase activity | 5 | 0.88 | 1.7e-03 |
| GO:0005057 | receptor signaling protein activity | 5 | 0.90 | 1.9e-03 |
| GO:0015631 | tubulin binding | 4 | 0.60 | 2.8e-03 |
| GO:0016879 | ligase activity, forming carbon-nitrogen bonds | 12 | 4.92 | 4.1e-03 |
| GO:0016874 | ligase activity | 15 | 6.91 | 4.2e-03 |
| GO:0003677 | DNA binding | 59 | 41.81 | 4.3e-03 |
| GO:0030508 | thiol-disulfide exchange intermediate activity | 6 | 1.64 | 5.9e-03 |
| GO:0016773 | phosphotransferase activity, alcohol group as acceptor | 33 | 20.86 | 6.5e-03 |
| GO:0004806 | triacylglycerol lipase activity | 4 | 0.79 | 7.5e-03 |
| GO:0004672 | protein kinase activity | 29 | 18.02 | 8.3e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009628 | response to abiotic stimulus | 67 | 29.88 | 3.2e-10 |
| GO:0042221 | response to chemical stimulus | 52 | 20.26 | 4.2e-10 |
| GO:0009719 | response to endogenous stimulus | 41 | 15.80 | 2.5e-08 |
| GO:0009725 | response to hormone stimulus | 32 | 11.14 | 9.6e-08 |
| GO:0050896 | response to stimulus | 76 | 43.94 | 1.3e-06 |
| GO:0009605 | response to external stimulus | 18 | 5.48 | 1.1e-05 |
| GO:0006914 | autophagy | 5 | 0.41 | 3.9e-05 |
| GO:0009733 | response to auxin stimulus | 15 | 4.60 | 6.3e-05 |
| GO:0000302 | response to reactive oxygen species | 5 | 0.45 | 6.8e-05 |
| GO:0046686 | response to cadmium ion | 7 | 1.06 | 8.3e-05 |
| GO:0009739 | response to gibberellic acid stimulus | 9 | 1.97 | 1.5e-04 |
| GO:0009723 | response to ethylene stimulus | 10 | 2.63 | 3.1e-04 |
| GO:0006950 | response to stress | 42 | 24.38 | 4.2e-04 |
| GO:0009737 | response to abscisic acid stimulus | 12 | 3.83 | 4.8e-04 |
| GO:0009056 | catabolism | 25 | 12.09 | 5.2e-04 |
| GO:0010038 | response to metal ion | 7 | 1.49 | 7.1e-04 |
| GO:0009753 | response to jasmonic acid stimulus | 9 | 2.42 | 7.3e-04 |
| GO:0044248 | cellular catabolism | 24 | 11.70 | 7.6e-04 |
| GO:0009755 | hormone-mediated signaling | 11 | 3.49 | 7.8e-04 |
| GO:0009414 | response to water deprivation | 8 | 2.17 | 1.5e-03 |
| GO:0009861 | jasmonic acid and ethylene-dependent systemic resistance | 9 | 2.72 | 1.6e-03 |
| GO:0007242 | intracellular signaling cascade | 18 | 8.22 | 1.7e-03 |
| GO:0010035 | response to inorganic substance | 7 | 1.74 | 1.8e-03 |
| GO:0050789 | regulation of biological process | 56 | 37.83 | 1.9e-03 |
| GO:0009751 | response to salicylic acid stimulus | 8 | 2.26 | 2.0e-03 |
| GO:0006970 | response to osmotic stress | 11 | 3.98 | 2.3e-03 |
| GO:0006979 | response to oxidative stress | 10 | 3.44 | 2.5e-03 |
| GO:0009415 | response to water | 8 | 2.35 | 2.5e-03 |
| GO:0007582 | physiological process | 245 | 216.80 | 2.8e-03 |
| GO:0048366 | leaf development | 6 | 1.47 | 3.5e-03 |
| GO:0048827 | phyllome development | 6 | 1.49 | 3.8e-03 |
| GO:0009991 | response to extracellular stimulus | 4 | 0.68 | 4.5e-03 |
| GO:0006800 | oxygen and reactive oxygen species metabolism | 10 | 3.76 | 4.6e-03 |
| GO:0009611 | response to wounding | 10 | 3.78 | 4.8e-03 |
| GO:0050791 | regulation of physiological process | 51 | 35.29 | 4.9e-03 |
| GO:0006351 | transcription, DNA-dependent | 30 | 18.22 | 5.3e-03 |
| GO:0008152 | metabolism | 196 | 170.41 | 5.8e-03 |
| GO:0006355 | regulation of transcription, DNA-dependent | 29 | 17.63 | 6.2e-03 |
| GO:0009743 | response to carbohydrate stimulus | 5 | 1.18 | 6.3e-03 |
| GO:0010033 | response to organic substance | 5 | 1.18 | 6.3e-03 |
| GO:0048367 | shoot development | 7 | 2.22 | 6.9e-03 |
| GO:0019222 | regulation of metabolism | 46 | 31.69 | 6.9e-03 |
| GO:0007154 | cell communication | 28 | 17.00 | 6.9e-03 |
| GO:0006350 | transcription | 46 | 31.85 | 7.5e-03 |
| GO:0051244 | regulation of cellular physiological process | 49 | 34.43 | 7.7e-03 |
| GO:0050875 | cellular physiological process | 220 | 195.27 | 7.8e-03 |
| GO:0045449 | regulation of transcription | 44 | 30.31 | 8.2e-03 |
| GO:0050794 | regulation of cellular process | 49 | 34.61 | 8.4e-03 |
| GO:0031323 | regulation of cellular metabolism | 45 | 31.40 | 9.3e-03 |
| GO:0019219 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 44 | 30.56 | 9.3e-03 |
| GO:0044237 | cellular metabolism | 174 | 150.61 | 9.4e-03 |
| GO:0009738 | abscisic acid mediated signaling | 4 | 0.84 | 9.5e-03 |
| GO:0009416 | response to light stimulus | 12 | 5.50 | 9.6e-03 |
| GO:0009987 | cellular process | 226 | 202.08 | 9.8e-03 |
| GO:0007568 | aging | 5 | 1.31 | 9.9e-03 |
| GO:0009651 | response to salt stress | 9 | 3.58 | 1.0e-02 |
|
list of genes (text format)
Cluster 7 - contains 207 genes
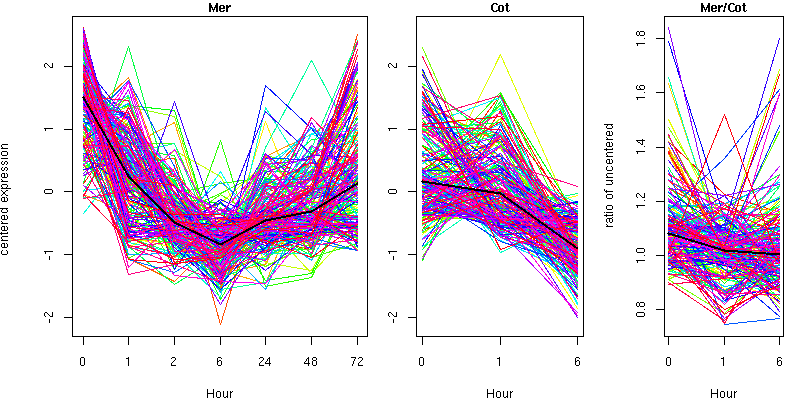
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003676 | nucleic acid binding | 41 | 25 | 0.00064 |
| GO:0030528 | transcription regulator activity | 28 | 15 | 0.00089 |
| GO:0003700 | transcription factor activity | 25 | 14 | 0.00210 |
| GO:0003677 | DNA binding | 29 | 18 | 0.00526 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009737 | response to abscisic acid stimulus | 8 | 1.6 | 0.00024 |
| GO:0009725 | response to hormone stimulus | 13 | 4.8 | 0.00102 |
| GO:0006350 | transcription | 26 | 13.6 | 0.00105 |
| GO:0051244 | regulation of cellular physiological process | 27 | 14.7 | 0.00151 |
| GO:0050794 | regulation of cellular process | 27 | 14.8 | 0.00163 |
| GO:0006970 | response to osmotic stress | 7 | 1.7 | 0.00164 |
| GO:0046907 | intracellular transport | 11 | 3.9 | 0.00178 |
| GO:0051649 | establishment of cellular localization | 11 | 3.9 | 0.00192 |
| GO:0051641 | cellular localization | 11 | 3.9 | 0.00196 |
| GO:0050791 | regulation of physiological process | 27 | 15.1 | 0.00215 |
| GO:0045449 | regulation of transcription | 24 | 12.9 | 0.00248 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 34 | 20.9 | 0.00273 |
| GO:0019219 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 24 | 13.1 | 0.00275 |
| GO:0050789 | regulation of biological process | 28 | 16.2 | 0.00285 |
| GO:0031323 | regulation of cellular metabolism | 24 | 13.4 | 0.00388 |
| GO:0019222 | regulation of metabolism | 24 | 13.5 | 0.00436 |
| GO:0015031 | protein transport | 8 | 2.8 | 0.00692 |
| GO:0006886 | intracellular protein transport | 7 | 2.2 | 0.00748 |
| GO:0045184 | establishment of protein localization | 8 | 2.9 | 0.00842 |
| GO:0008104 | protein localization | 8 | 2.9 | 0.00842 |
| GO:0007242 | intracellular signaling cascade | 9 | 3.5 | 0.00894 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0005634 | nucleus | 31 | 16 | 0.00042 |
| GO:0005622 | intracellular | 100 | 82 | 0.00536 |
|
list of genes (text format)
Cluster 8 - contains 537 genes
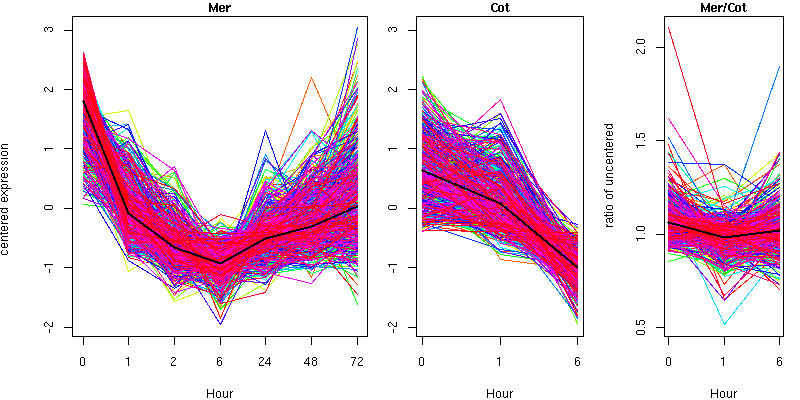
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0008270 | zinc ion binding | 40 | 20.97 | 7.5e-05 |
| GO:0003702 | RNA polymerase II transcription factor activity | 4 | 0.51 | 1.5e-03 |
| GO:0003723 | RNA binding | 20 | 10.00 | 2.7e-03 |
| GO:0003676 | nucleic acid binding | 86 | 64.66 | 3.2e-03 |
| GO:0008134 | transcription factor binding | 4 | 0.64 | 3.5e-03 |
| GO:0003682 | chromatin binding | 4 | 0.69 | 4.7e-03 |
| GO:0046914 | transition metal ion binding | 45 | 30.33 | 5.3e-03 |
| GO:0005515 | protein binding | 52 | 36.83 | 7.4e-03 |
| GO:0005488 | binding | 184 | 159.49 | 9.9e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0016192 | vesicle-mediated transport | 11 | 2.81 | 0.00012 |
| GO:0006352 | transcription initiation | 5 | 0.76 | 0.00086 |
| GO:0048193 | Golgi vesicle transport | 6 | 1.24 | 0.00142 |
| GO:0016049 | cell growth | 9 | 3.06 | 0.00368 |
| GO:0008361 | regulation of cell size | 9 | 3.17 | 0.00456 |
| GO:0048522 | positive regulation of cellular process | 5 | 1.19 | 0.00652 |
| GO:0051242 | positive regulation of cellular physiological process | 5 | 1.19 | 0.00652 |
|
list of genes (text format)
Cluster 9 - contains 457 genes
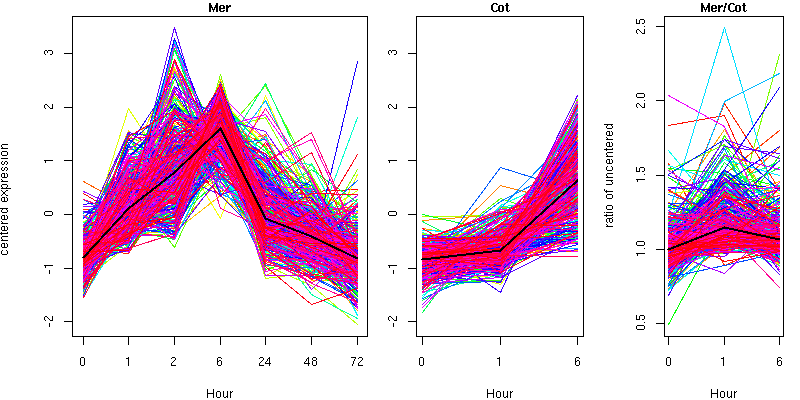
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016538 | cyclin-dependent protein kinase regulator activity | 8 | 0.48 | 1.3e-08 |
| GO:0019887 | protein kinase regulator activity | 8 | 0.75 | 5.7e-07 |
| GO:0019207 | kinase regulator activity | 8 | 0.77 | 7.3e-07 |
| GO:0003824 | catalytic activity | 183 | 143.10 | 2.7e-05 |
| GO:0016740 | transferase activity | 76 | 49.71 | 9.9e-05 |
| GO:0030234 | enzyme regulator activity | 15 | 4.89 | 1.3e-04 |
| GO:0004693 | cyclin-dependent protein kinase activity | 5 | 0.60 | 2.7e-04 |
| GO:0003777 | microtubule motor activity | 7 | 1.41 | 5.1e-04 |
| GO:0003774 | motor activity | 7 | 1.74 | 1.8e-03 |
| GO:0016772 | transferase activity, transferring phosphorus-containing groups | 44 | 29.63 | 5.5e-03 |
| GO:0017076 | purine nucleotide binding | 45 | 30.73 | 6.5e-03 |
| GO:0008168 | methyltransferase activity | 8 | 2.78 | 6.8e-03 |
| GO:0016759 | cellulose synthase activity | 4 | 0.77 | 7.1e-03 |
| GO:0004674 | protein serine/threonine kinase activity | 25 | 14.75 | 7.4e-03 |
| GO:0016741 | transferase activity, transferring one-carbon groups | 8 | 2.82 | 7.5e-03 |
| GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity | 5 | 1.28 | 8.9e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006334 | nucleosome assembly | 16 | 1.24 | 4.8e-14 |
| GO:0031497 | chromatin assembly | 17 | 1.49 | 6.7e-14 |
| GO:0006259 | DNA metabolism | 33 | 6.84 | 7.4e-14 |
| GO:0006323 | DNA packaging | 21 | 2.57 | 1.0e-13 |
| GO:0006325 | establishment and/or maintenance of chromatin architecture | 21 | 2.57 | 1.0e-13 |
| GO:0006333 | chromatin assembly or disassembly | 18 | 1.80 | 1.5e-13 |
| GO:0051276 | chromosome organization and biogenesis | 22 | 2.97 | 2.2e-13 |
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 21 | 2.73 | 3.5e-13 |
| GO:0006461 | protein complex assembly | 17 | 1.91 | 5.4e-12 |
| GO:0006996 | organelle organization and biogenesis | 37 | 11.21 | 2.1e-10 |
| GO:0007049 | cell cycle | 16 | 2.62 | 7.6e-09 |
| GO:0016043 | cell organization and biogenesis | 53 | 23.31 | 1.8e-08 |
| GO:0000074 | regulation of progression through cell cycle | 12 | 1.66 | 9.0e-08 |
| GO:0051726 | regulation of cell cycle | 12 | 1.66 | 9.0e-08 |
| GO:0009987 | cellular process | 247 | 198.16 | 5.3e-07 |
| GO:0006260 | DNA replication | 11 | 1.80 | 1.7e-06 |
| GO:0007582 | physiological process | 256 | 212.59 | 6.7e-06 |
| GO:0007017 | microtubule-based process | 10 | 1.91 | 2.0e-05 |
| GO:0006261 | DNA-dependent DNA replication | 7 | 0.89 | 2.5e-05 |
| GO:0050875 | cellular physiological process | 232 | 191.48 | 2.8e-05 |
| GO:0007010 | cytoskeleton organization and biogenesis | 13 | 3.51 | 5.3e-05 |
| GO:0043170 | macromolecule metabolism | 117 | 84.51 | 6.8e-05 |
| GO:0043283 | biopolymer metabolism | 75 | 50.39 | 2.5e-04 |
| GO:0007267 | cell-cell signaling | 4 | 0.42 | 7.1e-04 |
| GO:0007154 | cell communication | 31 | 16.67 | 7.1e-04 |
| GO:0007018 | microtubule-based movement | 6 | 1.20 | 1.2e-03 |
| GO:0044238 | primary metabolism | 163 | 134.61 | 1.6e-03 |
| GO:0007167 | enzyme linked receptor protein signaling pathway | 9 | 2.84 | 2.2e-03 |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 9 | 2.84 | 2.2e-03 |
| GO:0008152 | metabolism | 194 | 167.11 | 3.7e-03 |
| GO:0000278 | mitotic cell cycle | 4 | 0.64 | 3.7e-03 |
| GO:0007165 | signal transduction | 27 | 15.56 | 4.0e-03 |
| GO:0030705 | cytoskeleton-dependent intracellular transport | 6 | 1.53 | 4.3e-03 |
| GO:0007264 | small GTPase mediated signal transduction | 6 | 1.64 | 6.0e-03 |
| GO:0044237 | cellular metabolism | 172 | 147.68 | 6.8e-03 |
| GO:0046907 | intracellular transport | 17 | 8.86 | 8.2e-03 |
| GO:0019438 | aromatic compound biosynthesis | 8 | 2.89 | 8.5e-03 |
| GO:0051649 | establishment of cellular localization | 17 | 8.95 | 9.0e-03 |
| GO:0006725 | aromatic compound metabolism | 12 | 5.46 | 9.1e-03 |
| GO:0051641 | cellular localization | 17 | 8.97 | 9.2e-03 |
|
list of genes (text format)
Cluster 10 - contains 111 genes
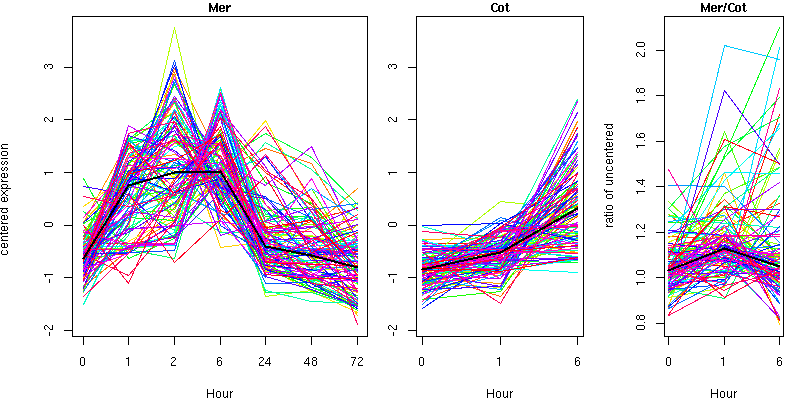
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005215 | transporter activity | 13 | 5.5 | 0.0035 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006260 | DNA replication | 4 | 0.41 | 0.00076 |
| GO:0044271 | nitrogen compound biosynthesis | 5 | 0.84 | 0.00154 |
| GO:0050875 | cellular physiological process | 58 | 43.60 | 0.00178 |
| GO:0007582 | physiological process | 62 | 48.41 | 0.00281 |
| GO:0019748 | secondary metabolism | 6 | 1.50 | 0.00390 |
| GO:0009266 | response to temperature stimulus | 5 | 1.05 | 0.00409 |
| GO:0009987 | cellular process | 58 | 45.13 | 0.00470 |
| GO:0006810 | transport | 18 | 9.37 | 0.00488 |
| GO:0051179 | localization | 18 | 9.43 | 0.00522 |
| GO:0051234 | establishment of localization | 18 | 9.43 | 0.00522 |
| GO:0006807 | nitrogen compound metabolism | 6 | 1.79 | 0.00924 |
|
list of genes (text format)
Cluster 11 - contains 501 genes
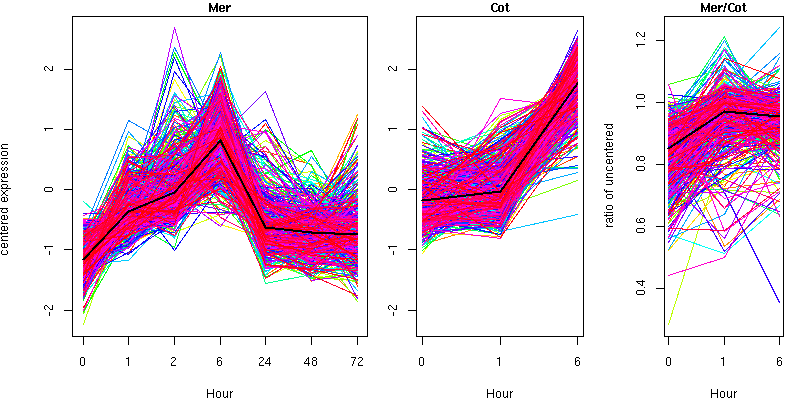
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003735 | structural constituent of ribosome | 90 | 7.18 | 4.0e-74 |
| GO:0005198 | structural molecule activity | 90 | 9.64 | 1.1e-61 |
| GO:0015450 | protein translocase activity | 7 | 0.99 | 5.0e-05 |
| GO:0003723 | RNA binding | 22 | 9.42 | 2.2e-04 |
| GO:0008565 | protein transporter activity | 9 | 2.65 | 1.4e-03 |
| GO:0015405 | P-P-bond-hydrolysis-driven transporter activity | 8 | 2.46 | 3.3e-03 |
| GO:0015399 | primary active transporter activity | 8 | 2.48 | 3.5e-03 |
| GO:0016835 | carbon-oxygen lyase activity | 8 | 2.63 | 4.9e-03 |
| GO:0016763 | transferase activity, transferring pentosyl groups | 4 | 0.70 | 4.9e-03 |
| GO:0016836 | hydro-lyase activity | 6 | 1.61 | 5.5e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0044249 | cellular biosynthesis | 144 | 41.84 | 1.2e-42 |
| GO:0006412 | protein biosynthesis | 100 | 20.93 | 9.7e-41 |
| GO:0009058 | biosynthesis | 147 | 45.96 | 8.0e-40 |
| GO:0009059 | macromolecule biosynthesis | 101 | 26.01 | 2.7e-33 |
| GO:0042254 | ribosome biogenesis and assembly | 39 | 3.20 | 1.0e-31 |
| GO:0007028 | cytoplasm organization and biogenesis | 39 | 3.20 | 1.0e-31 |
| GO:0007046 | ribosome biogenesis | 34 | 2.90 | 4.2e-27 |
| GO:0044267 | cellular protein metabolism | 129 | 63.79 | 5.1e-16 |
| GO:0019538 | protein metabolism | 132 | 66.16 | 5.9e-16 |
| GO:0006996 | organelle organization and biogenesis | 48 | 12.32 | 6.4e-16 |
| GO:0044260 | cellular macromolecule metabolism | 130 | 65.87 | 2.6e-15 |
| GO:0044237 | cellular metabolism | 241 | 162.30 | 1.1e-14 |
| GO:0043170 | macromolecule metabolism | 159 | 92.87 | 1.8e-13 |
| GO:0044238 | primary metabolism | 219 | 147.93 | 1.3e-12 |
| GO:0008152 | metabolism | 256 | 183.65 | 1.9e-12 |
| GO:0050875 | cellular physiological process | 275 | 210.44 | 3.3e-10 |
| GO:0009987 | cellular process | 278 | 217.78 | 4.1e-09 |
| GO:0007582 | physiological process | 293 | 233.64 | 5.2e-09 |
| GO:0016043 | cell organization and biogenesis | 57 | 25.62 | 1.1e-08 |
| GO:0044271 | nitrogen compound biosynthesis | 17 | 4.05 | 6.7e-07 |
| GO:0006520 | amino acid metabolism | 22 | 6.59 | 8.6e-07 |
| GO:0009309 | amine biosynthesis | 16 | 3.73 | 1.1e-06 |
| GO:0008652 | amino acid biosynthesis | 15 | 3.34 | 1.3e-06 |
| GO:0006519 | amino acid and derivative metabolism | 27 | 9.69 | 1.8e-06 |
| GO:0009308 | amine metabolism | 23 | 7.64 | 2.9e-06 |
| GO:0046148 | pigment biosynthesis | 9 | 1.34 | 6.6e-06 |
| GO:0006807 | nitrogen compound metabolism | 24 | 8.66 | 7.3e-06 |
| GO:0009112 | nucleobase metabolism | 6 | 0.51 | 8.1e-06 |
| GO:0006144 | purine base metabolism | 5 | 0.32 | 9.2e-06 |
| GO:0009113 | purine base biosynthesis | 4 | 0.17 | 1.2e-05 |
| GO:0046112 | nucleobase biosynthesis | 5 | 0.34 | 1.4e-05 |
| GO:0009075 | histidine family amino acid metabolism | 4 | 0.20 | 2.3e-05 |
| GO:0009076 | histidine family amino acid biosynthesis | 4 | 0.20 | 2.3e-05 |
| GO:0000105 | histidine biosynthesis | 4 | 0.20 | 2.3e-05 |
| GO:0006547 | histidine metabolism | 4 | 0.20 | 2.3e-05 |
| GO:0042440 | pigment metabolism | 9 | 1.56 | 2.4e-05 |
| GO:0019752 | carboxylic acid metabolism | 28 | 12.51 | 6.6e-05 |
| GO:0006082 | organic acid metabolism | 28 | 12.56 | 7.1e-05 |
| GO:0043037 | translation | 12 | 3.54 | 2.3e-04 |
| GO:0006414 | translational elongation | 5 | 0.59 | 2.4e-04 |
| GO:0006626 | protein targeting to mitochondrion | 4 | 0.41 | 6.5e-04 |
| GO:0009117 | nucleotide metabolism | 10 | 2.95 | 7.7e-04 |
| GO:0046483 | heterocycle metabolism | 11 | 3.59 | 9.7e-04 |
| GO:0006725 | aromatic compound metabolism | 15 | 6.00 | 1.1e-03 |
| GO:0009066 | aspartate family amino acid metabolism | 6 | 1.22 | 1.3e-03 |
| GO:0015979 | photosynthesis | 8 | 2.37 | 2.6e-03 |
| GO:0006779 | porphyrin biosynthesis | 5 | 1.00 | 3.1e-03 |
| GO:0006605 | protein targeting | 7 | 2.12 | 5.4e-03 |
| GO:0019748 | secondary metabolism | 15 | 7.22 | 6.3e-03 |
| GO:0006163 | purine nucleotide metabolism | 5 | 1.20 | 6.7e-03 |
| GO:0006164 | purine nucleotide biosynthesis | 5 | 1.20 | 6.7e-03 |
| GO:0009165 | nucleotide biosynthesis | 6 | 1.71 | 7.2e-03 |
| GO:0006778 | porphyrin metabolism | 5 | 1.22 | 7.3e-03 |
|
list of genes (text format)
Cluster 12 - contains 130 genes
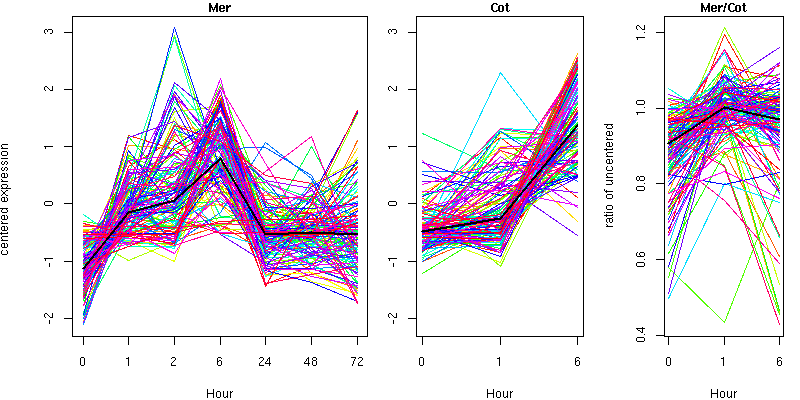
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003735 | structural constituent of ribosome | 7 | 1.8 | 0.0025 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0042254 | ribosome biogenesis and assembly | 5 | 0.79 | 0.0012 |
| GO:0007028 | cytoplasm organization and biogenesis | 5 | 0.79 | 0.0012 |
| GO:0009059 | macromolecule biosynthesis | 14 | 6.44 | 0.0049 |
| GO:0006412 | protein biosynthesis | 12 | 5.19 | 0.0057 |
| GO:0007046 | ribosome biogenesis | 4 | 0.72 | 0.0059 |
| GO:0044249 | cellular biosynthesis | 19 | 10.37 | 0.0068 |
| GO:0009058 | biosynthesis | 20 | 11.39 | 0.0087 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0044422 | organelle part | 20 | 8.00 | 0.00013 |
| GO:0044446 | intracellular organelle part | 20 | 8.00 | 0.00013 |
| GO:0030529 | ribonucleoprotein complex | 9 | 2.18 | 0.00035 |
| GO:0043228 | non-membrane-bound organelle | 11 | 3.93 | 0.00195 |
| GO:0043232 | intracellular non-membrane-bound organelle | 11 | 3.93 | 0.00195 |
| GO:0005840 | ribosome | 7 | 1.87 | 0.00279 |
| GO:0044445 | cytosolic part | 4 | 0.76 | 0.00713 |
|
list of genes (text format)
Cluster 13 - contains 152 genes
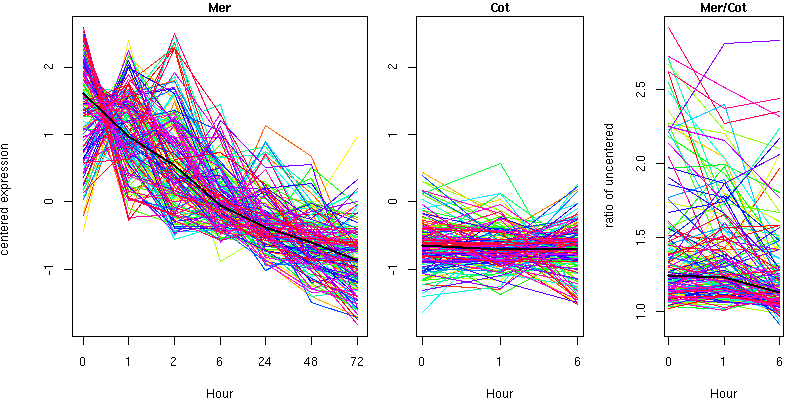
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005200 | structural constituent of cytoskeleton | 5 | 0.18 | 9.0e-07 |
| GO:0005525 | GTP binding | 8 | 1.40 | 8.6e-05 |
| GO:0019001 | guanyl nucleotide binding | 8 | 1.43 | 9.5e-05 |
| GO:0016829 | lyase activity | 9 | 2.09 | 2.6e-04 |
| GO:0016830 | carbon-carbon lyase activity | 5 | 0.57 | 2.8e-04 |
| GO:0003824 | catalytic activity | 63 | 47.24 | 3.5e-03 |
| GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 4 | 0.67 | 4.6e-03 |
| GO:0016853 | isomerase activity | 5 | 1.24 | 8.3e-03 |
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | 4 | 0.81 | 8.8e-03 |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | 7 | 2.36 | 9.6e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0007017 | microtubule-based process | 7 | 0.66 | 4.4e-06 |
| GO:0006996 | organelle organization and biogenesis | 14 | 3.88 | 3.6e-05 |
| GO:0006364 | rRNA processing | 4 | 0.27 | 1.5e-04 |
| GO:0051179 | localization | 29 | 14.35 | 1.7e-04 |
| GO:0051234 | establishment of localization | 29 | 14.35 | 1.7e-04 |
| GO:0016072 | rRNA metabolism | 4 | 0.28 | 1.8e-04 |
| GO:0007010 | cytoskeleton organization and biogenesis | 7 | 1.22 | 2.2e-04 |
| GO:0007264 | small GTPase mediated signal transduction | 5 | 0.57 | 2.6e-04 |
| GO:0006810 | transport | 28 | 14.25 | 3.6e-04 |
| GO:0016043 | cell organization and biogenesis | 19 | 8.08 | 4.4e-04 |
| GO:0005975 | carbohydrate metabolism | 14 | 5.11 | 6.1e-04 |
| GO:0051649 | establishment of cellular localization | 10 | 3.10 | 1.1e-03 |
| GO:0051641 | cellular localization | 10 | 3.11 | 1.2e-03 |
| GO:0007582 | physiological process | 91 | 73.67 | 2.0e-03 |
| GO:0046907 | intracellular transport | 9 | 3.07 | 3.8e-03 |
| GO:0044262 | cellular carbohydrate metabolism | 9 | 3.09 | 4.0e-03 |
| GO:0005996 | monosaccharide metabolism | 5 | 1.07 | 4.4e-03 |
| GO:0006091 | generation of precursor metabolites and energy | 13 | 5.67 | 4.5e-03 |
| GO:0050875 | cellular physiological process | 82 | 66.35 | 5.0e-03 |
| GO:0006732 | coenzyme metabolism | 5 | 1.12 | 5.3e-03 |
| GO:0009987 | cellular process | 84 | 68.67 | 5.8e-03 |
| GO:0015031 | protein transport | 7 | 2.21 | 6.8e-03 |
| GO:0045184 | establishment of protein localization | 7 | 2.28 | 8.2e-03 |
| GO:0008104 | protein localization | 7 | 2.28 | 8.2e-03 |
| GO:0006886 | intracellular protein transport | 6 | 1.78 | 9.2e-03 |
|
list of genes (text format)
Cluster 14 - contains 279 genes
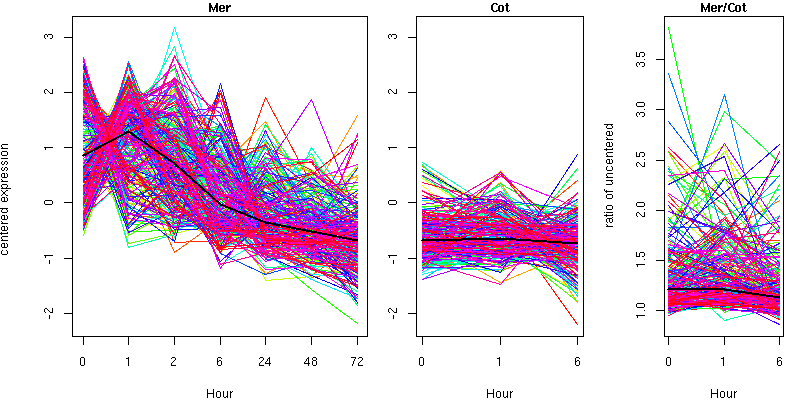
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0008092 | cytoskeletal protein binding | 5 | 0.98 | 0.003 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009725 | response to hormone stimulus | 17 | 6.62 | 0.00038 |
| GO:0043284 | biopolymer biosynthesis | 6 | 0.97 | 0.00042 |
| GO:0000271 | polysaccharide biosynthesis | 6 | 0.97 | 0.00042 |
| GO:0009628 | response to abiotic stimulus | 33 | 17.77 | 0.00042 |
| GO:0044264 | cellular polysaccharide metabolism | 6 | 1.14 | 0.00101 |
| GO:0042221 | response to chemical stimulus | 24 | 12.05 | 0.00105 |
| GO:0007017 | microtubule-based process | 6 | 1.16 | 0.00107 |
| GO:0005976 | polysaccharide metabolism | 6 | 1.17 | 0.00114 |
| GO:0009755 | hormone-mediated signaling | 8 | 2.07 | 0.00116 |
| GO:0007010 | cytoskeleton organization and biogenesis | 8 | 2.13 | 0.00137 |
| GO:0030243 | cellulose metabolism | 4 | 0.50 | 0.00149 |
| GO:0030244 | cellulose biosynthesis | 4 | 0.50 | 0.00149 |
| GO:0009664 | cell wall organization and biogenesis (sensu Magnoliophyta) | 6 | 1.24 | 0.00152 |
| GO:0042546 | cell wall biosynthesis | 5 | 0.85 | 0.00158 |
| GO:0009250 | glucan biosynthesis | 5 | 0.86 | 0.00170 |
| GO:0009832 | cell wall biosynthesis (sensu Magnoliophyta) | 4 | 0.54 | 0.00200 |
| GO:0008202 | steroid metabolism | 4 | 0.59 | 0.00285 |
| GO:0006073 | glucan metabolism | 5 | 1.04 | 0.00382 |
| GO:0016043 | cell organization and biogenesis | 25 | 14.13 | 0.00410 |
| GO:0006886 | intracellular protein transport | 9 | 3.12 | 0.00430 |
| GO:0016051 | carbohydrate biosynthesis | 8 | 2.84 | 0.00793 |
| GO:0009737 | response to abscisic acid stimulus | 7 | 2.27 | 0.00804 |
| GO:0009414 | response to water deprivation | 5 | 1.29 | 0.00962 |
|
list of genes (text format)
Cluster 15 - contains 192 genes
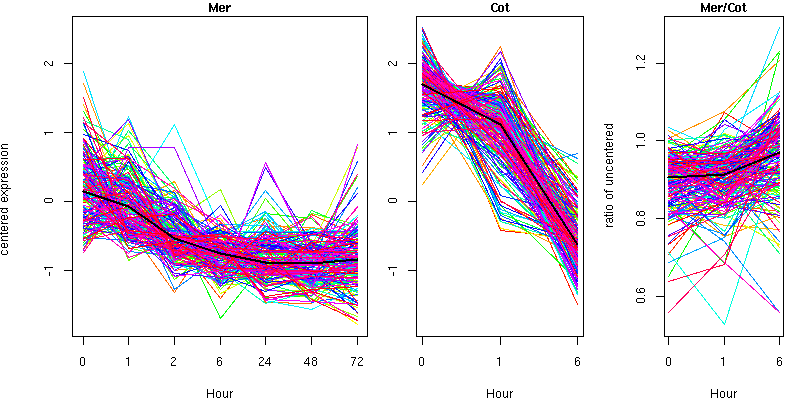
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 84 | 60.94 | 0.00023 |
| GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors | 4 | 0.43 | 0.00090 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006635 | fatty acid beta-oxidation | 4 | 0.14 | 8.6e-06 |
| GO:0007031 | peroxisome organization and biogenesis | 4 | 0.16 | 1.5e-05 |
| GO:0019395 | fatty acid oxidation | 4 | 0.17 | 1.9e-05 |
| GO:0051186 | cofactor metabolism | 10 | 1.94 | 2.6e-05 |
| GO:0005982 | starch metabolism | 4 | 0.22 | 6.3e-05 |
| GO:0005975 | carbohydrate metabolism | 17 | 6.09 | 1.3e-04 |
| GO:0044262 | cellular carbohydrate metabolism | 12 | 3.69 | 3.5e-04 |
| GO:0006732 | coenzyme metabolism | 7 | 1.33 | 3.9e-04 |
| GO:0051188 | cofactor biosynthesis | 6 | 1.04 | 6.1e-04 |
| GO:0044249 | cellular biosynthesis | 29 | 15.74 | 9.1e-04 |
| GO:0019752 | carboxylic acid metabolism | 13 | 4.71 | 9.2e-04 |
| GO:0009058 | biosynthesis | 31 | 17.29 | 9.2e-04 |
| GO:0006082 | organic acid metabolism | 13 | 4.73 | 9.5e-04 |
| GO:0006092 | main pathways of carbohydrate metabolism | 6 | 1.17 | 1.2e-03 |
| GO:0006725 | aromatic compound metabolism | 8 | 2.26 | 2.0e-03 |
| GO:0015980 | energy derivation by oxidation of organic compounds | 7 | 1.81 | 2.3e-03 |
| GO:0046483 | heterocycle metabolism | 6 | 1.35 | 2.4e-03 |
| GO:0043170 | macromolecule metabolism | 50 | 34.93 | 3.6e-03 |
| GO:0006767 | water-soluble vitamin metabolism | 4 | 0.63 | 3.7e-03 |
| GO:0008152 | metabolism | 86 | 69.08 | 5.0e-03 |
| GO:0009117 | nucleotide metabolism | 5 | 1.11 | 5.2e-03 |
| GO:0006073 | glucan metabolism | 4 | 0.71 | 5.5e-03 |
| GO:0006766 | vitamin metabolism | 4 | 0.72 | 6.0e-03 |
| GO:0044264 | cellular polysaccharide metabolism | 4 | 0.78 | 7.8e-03 |
| GO:0044248 | cellular catabolism | 11 | 4.74 | 8.3e-03 |
| GO:0006605 | protein targeting | 4 | 0.80 | 8.4e-03 |
| GO:0005976 | polysaccharide metabolism | 4 | 0.80 | 8.4e-03 |
| GO:0046164 | alcohol catabolism | 4 | 0.83 | 9.5e-03 |
|
list of genes (text format)
Cluster 16 - contains 305 genes
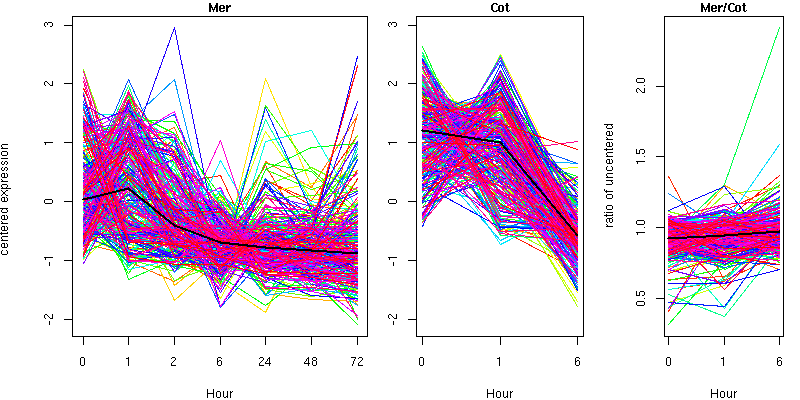
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 129 | 93.80 | 6.6e-06 |
| GO:0004177 | aminopeptidase activity | 4 | 0.26 | 1.1e-04 |
| GO:0008233 | peptidase activity | 17 | 6.53 | 3.3e-04 |
| GO:0016491 | oxidoreductase activity | 30 | 17.01 | 1.8e-03 |
| GO:0016829 | lyase activity | 11 | 4.15 | 3.2e-03 |
| GO:0008238 | exopeptidase activity | 5 | 1.05 | 4.1e-03 |
| GO:0015103 | inorganic anion transporter activity | 4 | 0.66 | 4.3e-03 |
| GO:0016830 | carbon-carbon lyase activity | 5 | 1.14 | 5.7e-03 |
| GO:0016787 | hydrolase activity | 45 | 31.01 | 6.6e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009657 | plastid organization and biogenesis | 7 | 0.70 | 5.8e-06 |
| GO:0019752 | carboxylic acid metabolism | 21 | 7.50 | 2.3e-05 |
| GO:0006082 | organic acid metabolism | 21 | 7.53 | 2.4e-05 |
| GO:0006732 | coenzyme metabolism | 10 | 2.12 | 5.5e-05 |
| GO:0009108 | coenzyme biosynthesis | 7 | 1.01 | 6.6e-05 |
| GO:0051186 | cofactor metabolism | 12 | 3.08 | 6.7e-05 |
| GO:0006508 | proteolysis | 20 | 8.13 | 2.1e-04 |
| GO:0006510 | ATP-dependent proteolysis | 4 | 0.34 | 3.2e-04 |
| GO:0009309 | amine biosynthesis | 9 | 2.24 | 4.2e-04 |
| GO:0008152 | metabolism | 136 | 110.02 | 7.6e-04 |
| GO:0044271 | nitrogen compound biosynthesis | 9 | 2.43 | 7.7e-04 |
| GO:0006807 | nitrogen compound metabolism | 14 | 5.19 | 7.7e-04 |
| GO:0006752 | group transfer coenzyme metabolism | 5 | 0.73 | 8.0e-04 |
| GO:0007582 | physiological process | 165 | 139.97 | 1.1e-03 |
| GO:0051188 | cofactor biosynthesis | 7 | 1.65 | 1.4e-03 |
| GO:0009308 | amine metabolism | 12 | 4.57 | 2.3e-03 |
| GO:0009069 | serine family amino acid metabolism | 4 | 0.58 | 2.7e-03 |
| GO:0008652 | amino acid biosynthesis | 7 | 2.00 | 4.0e-03 |
| GO:0009814 | defense response to pathogen, incompatible interaction | 8 | 2.67 | 5.6e-03 |
| GO:0006519 | amino acid and derivative metabolism | 13 | 5.80 | 5.8e-03 |
| GO:0006520 | amino acid metabolism | 10 | 3.95 | 6.6e-03 |
|
list of genes (text format)
Cluster 17 - contains 603 genes
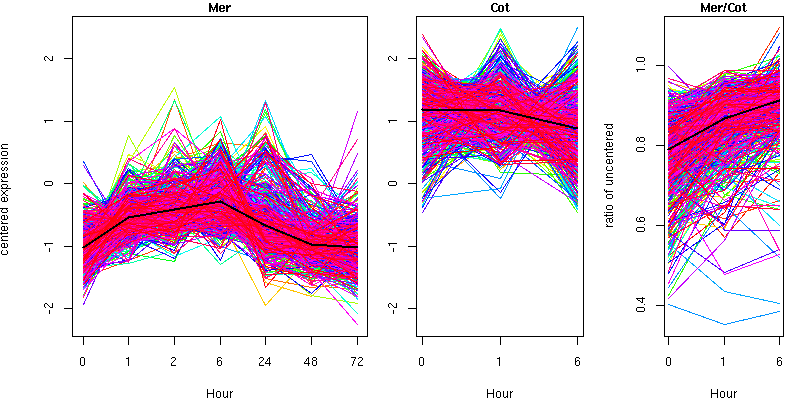
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0004812 | aminoacyl-tRNA ligase activity | 15 | 1.29 | 9.7e-13 |
| GO:0016875 | ligase activity, forming carbon-oxygen bonds | 15 | 1.29 | 9.7e-13 |
| GO:0016876 | ligase activity, forming aminoacyl-tRNA and related compounds | 15 | 1.29 | 9.7e-13 |
| GO:0005198 | structural molecule activity | 38 | 11.20 | 5.3e-11 |
| GO:0003735 | structural constituent of ribosome | 32 | 8.34 | 8.2e-11 |
| GO:0016853 | isomerase activity | 20 | 4.79 | 7.2e-08 |
| GO:0008462 | endopeptidase Clp activity | 5 | 0.25 | 1.9e-06 |
| GO:0016859 | cis-trans isomerase activity | 9 | 1.48 | 1.5e-05 |
| GO:0016874 | ligase activity | 22 | 8.37 | 3.9e-05 |
| GO:0003723 | RNA binding | 26 | 10.95 | 4.6e-05 |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 8 | 1.46 | 9.1e-05 |
| GO:0003824 | catalytic activity | 223 | 181.79 | 9.8e-05 |
| GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor | 4 | 0.28 | 1.1e-04 |
| GO:0008237 | metallopeptidase activity | 9 | 1.99 | 1.6e-04 |
| GO:0008483 | transaminase activity | 7 | 1.23 | 2.0e-04 |
| GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 10 | 2.58 | 2.5e-04 |
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | 11 | 3.11 | 2.8e-04 |
| GO:0016769 | transferase activity, transferring nitrogenous groups | 7 | 1.32 | 3.1e-04 |
| GO:0004222 | metalloendopeptidase activity | 6 | 1.06 | 6.0e-04 |
| GO:0016866 | intramolecular transferase activity | 6 | 1.09 | 7.0e-04 |
| GO:0008236 | serine-type peptidase activity | 12 | 4.12 | 9.0e-04 |
| GO:0016741 | transferase activity, transferring one-carbon groups | 11 | 3.58 | 9.5e-04 |
| GO:0004252 | serine-type endopeptidase activity | 9 | 2.58 | 1.1e-03 |
| GO:0016836 | hydro-lyase activity | 7 | 1.88 | 2.6e-03 |
| GO:0008168 | methyltransferase activity | 10 | 3.53 | 2.9e-03 |
| GO:0005527 | macrolide binding | 4 | 0.62 | 3.0e-03 |
| GO:0005528 | FK506 binding | 4 | 0.62 | 3.0e-03 |
| GO:0016835 | carbon-oxygen lyase activity | 9 | 3.05 | 3.5e-03 |
| GO:0008144 | drug binding | 4 | 0.67 | 4.1e-03 |
| GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 7 | 2.07 | 4.6e-03 |
| GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides | 5 | 1.12 | 5.0e-03 |
| GO:0004175 | endopeptidase activity | 16 | 7.78 | 5.4e-03 |
| GO:0008233 | peptidase activity | 22 | 12.66 | 8.9e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009058 | biosynthesis | 121 | 52.48 | 4.9e-19 |
| GO:0044249 | cellular biosynthesis | 113 | 47.77 | 1.5e-18 |
| GO:0006399 | tRNA metabolism | 23 | 2.26 | 2.2e-17 |
| GO:0006520 | amino acid metabolism | 39 | 7.52 | 2.6e-17 |
| GO:0019752 | carboxylic acid metabolism | 52 | 14.29 | 6.5e-16 |
| GO:0006082 | organic acid metabolism | 52 | 14.35 | 7.6e-16 |
| GO:0006807 | nitrogen compound metabolism | 42 | 9.89 | 2.2e-15 |
| GO:0009308 | amine metabolism | 39 | 8.72 | 4.2e-15 |
| GO:0006418 | tRNA aminoacylation for protein translation | 16 | 1.36 | 1.5e-13 |
| GO:0043038 | amino acid activation | 16 | 1.36 | 1.5e-13 |
| GO:0043039 | tRNA aminoacylation | 16 | 1.36 | 1.5e-13 |
| GO:0043037 | translation | 25 | 4.04 | 2.6e-13 |
| GO:0006519 | amino acid and derivative metabolism | 40 | 11.06 | 2.1e-12 |
| GO:0046483 | heterocycle metabolism | 22 | 4.09 | 1.3e-10 |
| GO:0016070 | RNA metabolism | 30 | 7.97 | 4.9e-10 |
| GO:0006779 | porphyrin biosynthesis | 12 | 1.14 | 7.3e-10 |
| GO:0006412 | protein biosynthesis | 57 | 23.90 | 1.0e-09 |
| GO:0009790 | embryonic development | 27 | 6.96 | 1.9e-09 |
| GO:0051186 | cofactor metabolism | 24 | 5.88 | 5.6e-09 |
| GO:0006778 | porphyrin metabolism | 12 | 1.39 | 8.9e-09 |
| GO:0009793 | embryonic development (sensu Magnoliophyta) | 25 | 6.52 | 9.7e-09 |
| GO:0009059 | macromolecule biosynthesis | 63 | 29.70 | 1.2e-08 |
| GO:0048316 | seed development | 25 | 7.02 | 4.3e-08 |
| GO:0008652 | amino acid biosynthesis | 18 | 3.82 | 5.0e-08 |
| GO:0048608 | reproductive structure development | 25 | 7.10 | 5.4e-08 |
| GO:0009657 | plastid organization and biogenesis | 11 | 1.34 | 6.3e-08 |
| GO:0009658 | chloroplast organization and biogenesis | 9 | 0.84 | 8.0e-08 |
| GO:0051188 | cofactor biosynthesis | 16 | 3.15 | 9.6e-08 |
| GO:0009309 | amine biosynthesis | 18 | 4.26 | 2.7e-07 |
| GO:0044271 | nitrogen compound biosynthesis | 18 | 4.62 | 9.2e-07 |
| GO:0048481 | ovule development | 7 | 0.61 | 1.5e-06 |
| GO:0000003 | reproduction | 26 | 9.22 | 2.2e-06 |
| GO:0046148 | pigment biosynthesis | 10 | 1.53 | 2.4e-06 |
| GO:0042440 | pigment metabolism | 10 | 1.78 | 1.0e-05 |
| GO:0048440 | carpel development | 7 | 0.81 | 1.1e-05 |
| GO:0048467 | gynoecium development | 7 | 0.89 | 2.3e-05 |
| GO:0008152 | metabolism | 255 | 209.71 | 2.4e-05 |
| GO:0006414 | translational elongation | 6 | 0.67 | 4.0e-05 |
| GO:0044237 | cellular metabolism | 227 | 185.33 | 7.1e-05 |
| GO:0008033 | tRNA processing | 6 | 0.81 | 1.2e-04 |
| GO:0048438 | floral whorl development | 7 | 1.17 | 1.4e-04 |
| GO:0009072 | aromatic amino acid family metabolism | 7 | 1.23 | 2.0e-04 |
| GO:0007275 | development | 45 | 25.80 | 2.1e-04 |
| GO:0019538 | protein metabolism | 105 | 75.55 | 2.2e-04 |
| GO:0043170 | macromolecule metabolism | 139 | 106.05 | 2.4e-04 |
| GO:0006457 | protein folding | 15 | 5.26 | 2.7e-04 |
| GO:0015995 | chlorophyll biosynthesis | 5 | 0.61 | 2.9e-04 |
| GO:0042254 | ribosome biogenesis and assembly | 12 | 3.65 | 3.0e-04 |
| GO:0007028 | cytoplasm organization and biogenesis | 12 | 3.65 | 3.0e-04 |
| GO:0044267 | cellular protein metabolism | 101 | 72.85 | 3.3e-04 |
| GO:0006510 | ATP-dependent proteolysis | 5 | 0.64 | 3.7e-04 |
| GO:0044260 | cellular macromolecule metabolism | 103 | 75.21 | 4.4e-04 |
| GO:0009067 | aspartate family amino acid biosynthesis | 5 | 0.67 | 4.5e-04 |
| GO:0007046 | ribosome biogenesis | 11 | 3.31 | 4.9e-04 |
| GO:0009073 | aromatic amino acid family biosynthesis | 6 | 1.06 | 5.9e-04 |
| GO:0048569 | post-embryonic organ development | 7 | 1.50 | 7.1e-04 |
| GO:0048437 | floral organ development | 7 | 1.50 | 7.1e-04 |
| GO:0045036 | protein targeting to chloroplast | 4 | 0.45 | 8.3e-04 |
| GO:0015994 | chlorophyll metabolism | 5 | 0.78 | 9.5e-04 |
| GO:0016117 | carotenoid biosynthesis | 4 | 0.47 | 1.1e-03 |
| GO:0016109 | tetraterpenoid biosynthesis | 4 | 0.47 | 1.1e-03 |
| GO:0016116 | carotenoid metabolism | 4 | 0.58 | 2.4e-03 |
| GO:0016108 | tetraterpenoid metabolism | 4 | 0.58 | 2.4e-03 |
| GO:0006568 | tryptophan metabolism | 4 | 0.58 | 2.4e-03 |
| GO:0006586 | indolalkylamine metabolism | 4 | 0.58 | 2.4e-03 |
| GO:0009066 | aspartate family amino acid metabolism | 6 | 1.39 | 2.6e-03 |
| GO:0044238 | primary metabolism | 199 | 168.93 | 2.6e-03 |
| GO:0042430 | indole and derivative metabolism | 4 | 0.70 | 4.7e-03 |
| GO:0042434 | indole derivative metabolism | 4 | 0.70 | 4.7e-03 |
| GO:0042435 | indole derivative biosynthesis | 4 | 0.70 | 4.7e-03 |
| GO:0006766 | vitamin metabolism | 7 | 2.20 | 6.4e-03 |
| GO:0006839 | mitochondrial transport | 5 | 1.20 | 6.6e-03 |
| GO:0006396 | RNA processing | 12 | 5.29 | 7.1e-03 |
| GO:0009110 | vitamin biosynthesis | 5 | 1.28 | 8.8e-03 |
| GO:0008610 | lipid biosynthesis | 14 | 6.91 | 1.0e-02 |
|
list of genes (text format)
Cluster 18 - contains 127 genes
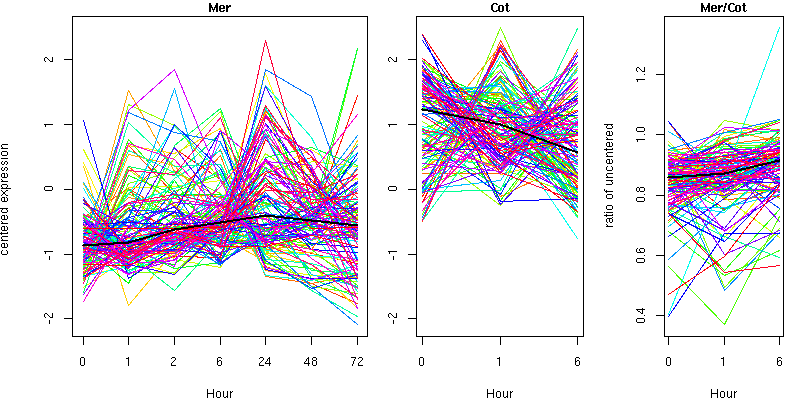
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 55 | 39.0 | 0.0014 |
| GO:0016491 | oxidoreductase activity | 15 | 7.1 | 0.0046 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006520 | amino acid metabolism | 10 | 1.56 | 3.7e-06 |
| GO:0006807 | nitrogen compound metabolism | 11 | 2.05 | 6.5e-06 |
| GO:0009308 | amine metabolism | 10 | 1.81 | 1.3e-05 |
| GO:0006519 | amino acid and derivative metabolism | 10 | 2.29 | 1.0e-04 |
| GO:0019752 | carboxylic acid metabolism | 10 | 2.96 | 7.6e-04 |
| GO:0006082 | organic acid metabolism | 10 | 2.97 | 7.9e-04 |
| GO:0044271 | nitrogen compound biosynthesis | 5 | 0.96 | 2.8e-03 |
| GO:0008652 | amino acid biosynthesis | 4 | 0.79 | 8.2e-03 |
| GO:0016052 | carbohydrate catabolism | 4 | 0.80 | 8.4e-03 |
| GO:0044275 | cellular carbohydrate catabolism | 4 | 0.80 | 8.4e-03 |
| GO:0008152 | metabolism | 56 | 43.43 | 8.6e-03 |
|
list of genes (text format)
Cluster 19 - contains 176 genes
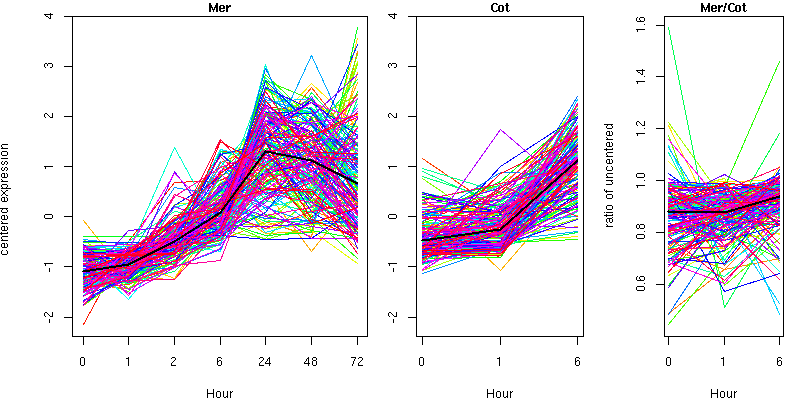
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016491 | oxidoreductase activity | 31 | 9.56 | 4.8e-09 |
| GO:0003824 | catalytic activity | 78 | 52.72 | 1.9e-05 |
| GO:0016829 | lyase activity | 10 | 2.33 | 1.2e-04 |
| GO:0009055 | electron carrier activity | 7 | 1.84 | 2.6e-03 |
| GO:0030508 | thiol-disulfide exchange intermediate activity | 4 | 0.58 | 2.7e-03 |
| GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 4 | 0.60 | 3.1e-03 |
| GO:0015036 | disulfide oxidoreductase activity | 5 | 1.03 | 3.8e-03 |
| GO:0016830 | carbon-carbon lyase activity | 4 | 0.64 | 3.9e-03 |
| GO:0015035 | protein disulfide oxidoreductase activity | 4 | 0.66 | 4.3e-03 |
|
list of genes (text format)
Cluster 20 - contains 264 genes
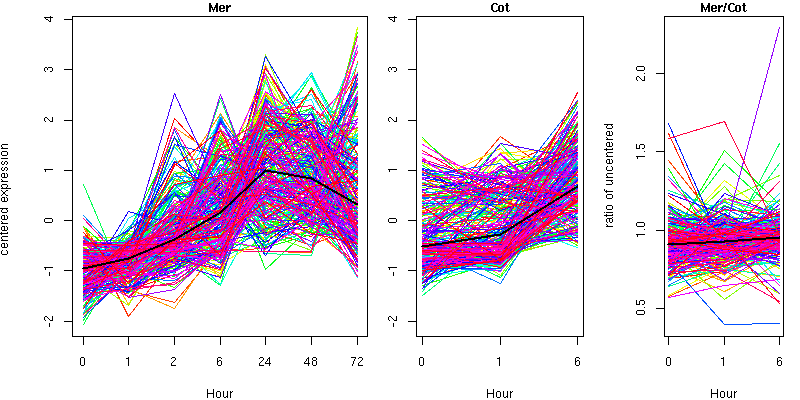
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016491 | oxidoreductase activity | 39 | 14.40 | 1.2e-08 |
| GO:0003824 | catalytic activity | 115 | 79.43 | 9.4e-07 |
| GO:0016168 | chlorophyll binding | 5 | 0.28 | 7.4e-06 |
| GO:0009055 | electron carrier activity | 12 | 2.78 | 2.4e-05 |
| GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors | 4 | 0.40 | 6.8e-04 |
| GO:0016779 | nucleotidyltransferase activity | 7 | 1.77 | 2.1e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0015979 | photosynthesis | 13 | 1.19 | 2.0e-10 |
| GO:0019684 | photosynthesis, light reaction | 9 | 0.73 | 4.1e-08 |
| GO:0009765 | photosynthesis, light harvesting | 6 | 0.28 | 2.8e-07 |
| GO:0009642 | response to light intensity | 5 | 0.31 | 1.2e-05 |
| GO:0006091 | generation of precursor metabolites and energy | 24 | 9.07 | 1.4e-05 |
| GO:0007582 | physiological process | 149 | 117.87 | 1.5e-05 |
| GO:0009657 | plastid organization and biogenesis | 6 | 0.59 | 2.6e-05 |
| GO:0009628 | response to abiotic stimulus | 34 | 16.25 | 3.3e-05 |
| GO:0009069 | serine family amino acid metabolism | 5 | 0.49 | 1.3e-04 |
| GO:0008152 | metabolism | 120 | 92.65 | 1.4e-04 |
| GO:0009809 | lignin biosynthesis | 4 | 0.30 | 2.0e-04 |
| GO:0009808 | lignin metabolism | 4 | 0.32 | 2.7e-04 |
| GO:0019748 | secondary metabolism | 12 | 3.64 | 3.2e-04 |
| GO:0050896 | response to stimulus | 41 | 23.89 | 3.9e-04 |
| GO:0006519 | amino acid and derivative metabolism | 14 | 4.89 | 4.2e-04 |
| GO:0006807 | nitrogen compound metabolism | 13 | 4.37 | 4.8e-04 |
| GO:0042221 | response to chemical stimulus | 23 | 11.02 | 7.0e-04 |
| GO:0009416 | response to light stimulus | 10 | 2.99 | 8.9e-04 |
| GO:0009314 | response to radiation | 10 | 3.03 | 9.7e-04 |
| GO:0009308 | amine metabolism | 11 | 3.85 | 1.8e-03 |
| GO:0006520 | amino acid metabolism | 10 | 3.32 | 1.9e-03 |
| GO:0050875 | cellular physiological process | 128 | 106.17 | 2.0e-03 |
| GO:0009110 | vitamin biosynthesis | 4 | 0.57 | 2.4e-03 |
| GO:0044237 | cellular metabolism | 102 | 81.88 | 3.4e-03 |
| GO:0006725 | aromatic compound metabolism | 9 | 3.03 | 3.5e-03 |
| GO:0006996 | organelle organization and biogenesis | 14 | 6.22 | 4.0e-03 |
| GO:0044271 | nitrogen compound biosynthesis | 7 | 2.04 | 4.5e-03 |
| GO:0019438 | aromatic compound biosynthesis | 6 | 1.60 | 5.5e-03 |
| GO:0009987 | cellular process | 129 | 109.87 | 6.1e-03 |
| GO:0008652 | amino acid biosynthesis | 6 | 1.69 | 7.0e-03 |
| GO:0010038 | response to metal ion | 4 | 0.81 | 8.9e-03 |
|
list of genes (text format)




















