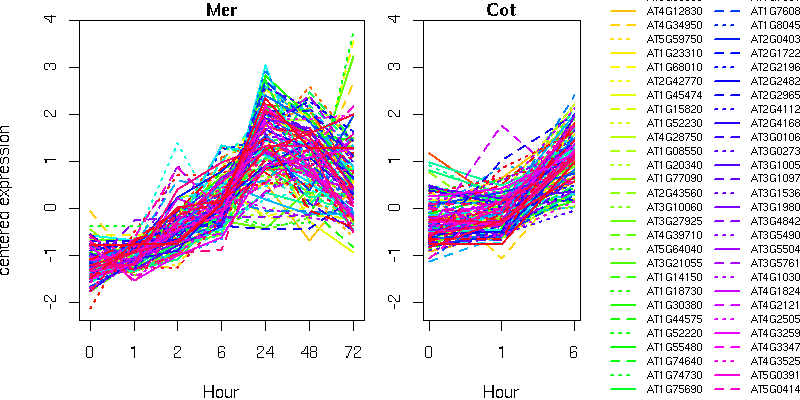
NA
NA
PMDH2 (PEROXISOMAL NAD-MALATE DEHYDROGENASE 2)
NA
FAD5 (FATTY ACID DESATURASE 5); oxidoreductase
NA
PGI1 (CHLOROPLASTIC PHOSPHOGLUCOSE ISOMERASE)
NA
NA
NA
NA
AGL79 (AGAMOUS-LIKE 79)
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G03055.1); similar to hypothetical protein LOC_Os11g37650 [Oryza sativa (japonica cultivar-group)] (GB:ABA94460.1); similar to Os08g0114100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001060847.1)
NA
NA
NA
NA
NA
GGT1 (ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE 1)
NA
peroxisomal membrane 22 kDa family protein
LHCA5 (Photosystem I light harvesting complex gene 5)
LHCB6 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding
photosystem I reaction center subunit VI, chloroplast, putative / PSI-H, putative (PSAH2)
PSAE-1 (PSA E1 KNOCKOUT)
NPQ1 (NON-PHOTOCHEMICAL QUENCHING 1)
NA
thylakoid lumenal 29.8 kDa protein
immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein
NA
NA
NA
PSAN (photosystem I reaction center subunit PSI-N); calmodulin binding
NA
oxygen evolving enhancer 3 (PsbQ) family protein
similar to Os02g0744000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001048099.1)
PSAK (PHOTOSYSTEM I SUBUNIT K)
NPQ4 (NONPHOTOCHEMICAL QUENCHING)
similar to TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA) [Arabidopsis thaliana] (TAIR:AT2G46820.2); similar to P0460E08.25 [Oryza sativa (japonica cultivar-group)] (GB:BAB61215.1); similar to Os01g0761000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001044320.1)
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G08050.1); similar to Os12g0575000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001067105.1); similar to Os01g0585300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001043425.1); contains InterPro domain Protein of unknown function DUF1118; (InterPro:IPR009500)
NA
NA
flavin reductase-related
RCA (RUBISCO ACTIVASE)
rhodanese-like domain-containing protein
NA
similar to unnamed protein product [Ostreococcus tauri] (GB:CAL56506.1); similar to Os01g0805200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001044558.1)
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G11960.1); similar to Os03g0857400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001051955.1); similar to Os08g0526300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001062297.1); similar to hypothetical protein OSJNBa0003M24.13 [Oryza sativa (japonica cultivar-group)] (GB:AAQ56398.1); contains domain no description (G3D.2.20.25.10); contains domain Zinc beta-ribbon (SSF57783)
NA
NA
NA
STN7 (STT7 HOMOLOG STN7); kinase/ protein kinase
NFU3 (NFU domain protein 3)
PSAO (photosystem I subunit O)
NA
NA
NA
NADK2 (NAD kinase 2); NAD+ kinase/ calmodulin binding
NA
similar to similar to Zn-dependent hydrolases of the beta-lactamase fold [Crocosphaera watsonii WH 8501] (GB:ZP_00516888.1); similar to Os12g0641300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001067402.1); contains domain no description (G3D.3.60.15.10); contains domain Metallo-hydrolase/oxidoreductase (SSF56281)
NA
NA
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57345.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:AAO19365.1); contains InterPro domain Protein of unknown function DUF1517; (InterPro:IPR010903)
amine oxidase family
NA
ISPF (Homolog of E. coli ispF (isoprenoids F)); 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
NA
UbiE/COQ5 methyltransferase family protein
NA
NA
NA
NA
protein kinase, putative
NA
TIC55 (TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 55); oxidoreductase
inorganic phosphate transporter, putative
NA
NA
similar to unknown [Brassica rapa] (GB:ABC41272.1)
ATF1/TRXF1 (THIOREDOXIN F-TYPE 1); thiol-disulfide exchange intermediate
NA
haloacid dehalogenase-like hydrolase family protein
NA
NA
haloacid dehalogenase-like hydrolase family protein
CXIP1 (CAX INTERACTING PROTEIN 1)
In2-1 protein, putative
adenylosuccinate synthetase (ADSS)
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04300.1); similar to Os04g0445200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001052900.1); similar to Cupin, RmlC-type [Medicago truncatula] (GB:ABE82642.1); similar to OSIGBa0140O07.4 [Oryza sativa (indica cultivar-group)] (GB:CAH67236.1); contains InterPro domain Protein of unknown function DUF861, cupin_3; (InterPro:IPR008579); contains InterPro domain AraC protein, arabinose-binding/dimerisation; (InterPro:IPR003313)
NA
Identical to UPF0085 protein At4g21210 [Arabidopsis Thaliana] (GB:O49562); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01200.1); similar to putative ATP/GTP-binding protein [Oryza sativa (japonica cultivar-group)] (GB:BAC45149.1); contains InterPro domain Protein of unknown function DUF299; (InterPro:IPR005177)
ACP4 (ACYL CARRIER PROTEIN 4)
electron carrier
HDA14 (histone deacetylase 14); histone deacetylase
NA
NA
NA
FTRA2 (ferredoxin/thioredoxin reductase subunit A (variable subunit) 2); ferredoxin:thioredoxin reductase
NA
NA
NA
NA
rhomboid family protein
hydrolase, alpha/beta fold family protein
CbbY protein-related
NA
NA
integral membrane HPP family protein
fructose-bisphosphate aldolase
fructose-bisphosphate aldolase, putative
CP12-2
60S ribosomal protein L7 (RPL7A)
list of genes (text format)