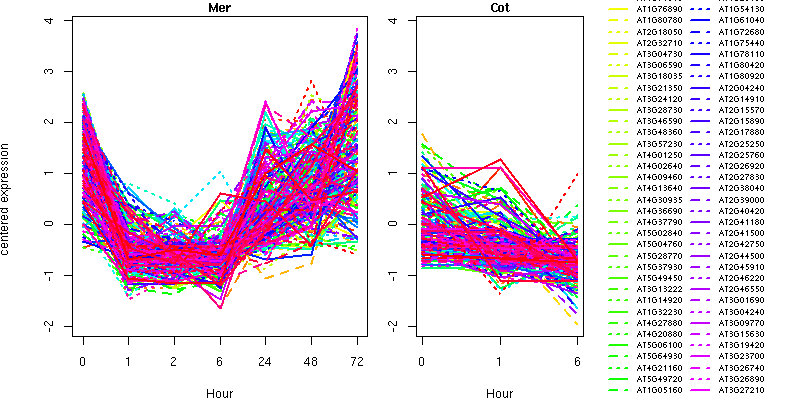
NA
AP2 domain-containing transcription factor, putative
RAP2.4 (related to AP2 4); DNA binding / transcription factor
WRKY15 (WRKY DNA-binding protein 15); transcription factor
ATHB-15 (INCURVATA 4); DNA binding / transcription factor
SIK1 (ERINE/THREONINE KINASE 1); kinase
casein kinase, putative
NA
WRKY40 (WRKY DNA-binding protein 40); transcription factor
WRKY25 (WRKY DNA-binding protein 25); transcription factor
GRF9 (General regulatory factor 9); protein phosphorylated amino acid binding
PIF4 (PHYTOCHROME INTERACTING FACTOR 4); DNA binding / transcription factor
NA
U1-70K (SPLICEOSOMAL PROTEIN U1A); RNA binding
NA
ankyrin protein kinase, putative
protein phosphatase 2C PPH1 / PP2C PPH1 (PPH1)
NA
NA
GRF6 (GF14 LAMBDA); protein phosphorylated amino acid binding
E2F1; transcription factor
NA
NA
SHY2 (SHORT HYPOCOTYL 2); transcription factor
NF-X1 type zinc finger family protein
NA
NA
myb family transcription factor
AP2 domain-containing transcription factor, putative
NA
NA
NA
RAV2 (REGULATOR OF THE ATPASE OF THE VACUOLAR MEMBRANE); DNA binding / transcription factor
NA
TRFL6 (TRF-LIKE 6); DNA binding / transcription factor
NA
NA
CCR4-NOT transcription complex protein, putative
NA
KRP4 (KIP-RELATED PROTEIN 4)
IAA16 (indoleacetic acid-induced protein 16); transcription factor
NA
HON4; DNA binding
NA
myb family transcription factor
ATHMG (HIGH MOBILITY GROUP, STRUCTURE-SPECIFIC RECOGNITION PROTEIN 1); transcription factor
TRFL1 (TRF-LIKE 1); DNA binding
BT2 (BTB and TAZ domain protein 2); protein binding / transcription regulator
NA
NA
BZO2H1 (basic leucine zipper O2 homolog 1); DNA binding / transcription factor
NA
UNE16 (unfertilized embryo sac 16)
NA
ATU2AF65A; RNA binding
NA
DNA binding / transcription factor
NA
BZO2H3 (basic leucine zipper O2 homolog 3); DNA binding / transcription factor
NA
bZIP family transcription factor
NA
GAI (GA INSENSITIVE); transcription factor
RCD1 (RADICAL-INDUCED CELL DEATH1)
seven in absentia (SINA) family protein
ethylene-responsive nuclear protein / ethylene-regulated nuclear protein (ERT2)
ATMYB33/MYB33 (myb domain protein 33)
NA
ZAC (ARF-GAP DOMAIN 12); ARF GTPase activator
KOR1 (KORRIGAN); hydrolase, hydrolyzing O-glycosyl compounds
CYP88A3 (ENT-KAURENOIC ACID HYDROXYLASE 1); oxygen binding
(AT)SRC2/SRC2 (SOYBEAN GENE REGULATED BY COLD-2); protein binding
HMG1 (3-HYDROXY-3-METHYLGLUTARYL COA REDUCTASE)
NA
FAD2 (FATTY ACID DESATURASE 2)
NA
NA
PDS1 (PHYTOENE DESATURATION 1)
CAT3 (CATALASE 3); catalase
ALDH6B2 (Aldehyde dehydrogenase 6B2); 3-chloroallyl aldehyde dehydrogenase
ATTIM17-2 (Arabidopsis thaliana translocase inner membrane subunit 17-2); protein translocase
NA
NA
ERD5 (EARLY RESPONSIVE TO DEHYDRATION 5, PROLINE OXIDASE); proline dehydrogenase
MCCB (3-METHYLCROTONYL-COA CARBOXYLASE); biotin carboxylase
CAT2 (CATALASE 2); catalase
NA
NA
NA
AtATG18g (Arabidopsis thaliana homolog of yeast autophagy 18 (ATG18) g)
ATPP2-A10 (Phloem protein 2-A10)
glycosyl transferase family 1 protein
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22110.1); similar to hypothetical protein [Oryza sativa (japonica cultivar-group)] (GB:BAD28434.1)
copine-related
NA
NA
NA
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G04090.2); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD10374.1); similar to Os02g0137100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001045827.1); contains InterPro domain Conserved hypothetical protein 1589, plant; (InterPro:IPR006476)
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G04860.1); similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABF97907.1); similar to conserved hypothetical protein [Medicago truncatula] (GB:ABE84189.1); similar to Os03g0649000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001050782.1)
auxin-responsive family protein
NA
UVR3 (UV REPAIR DEFECTIVE 4)
NA
SAC3/GANP family protein
shaggy-related protein kinase beta / ASK-beta (ASK2)
dehydration-responsive family protein
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30050.1); similar to Os05g0315200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055171.1); similar to Os11g0118800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001065600.1); similar to expressed protein-like protein [Sorghum bicolor] (GB:AAO16693.1)
NA
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G45210.1); similar to Protein of unknown function DUF584 [Medicago truncatula] (GB:ABE80969.1); contains InterPro domain Protein of unknown function DUF584; (InterPro:IPR007608)
NA
transcription initiation factor IID (TFIID) subunit A family protein
ACX4 (ACYL-COA OXIDASE 4); oxidoreductase
NA
similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABF94264.1)
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G40400.2); similar to Trimeric LpxA-like [Medicago truncatula] (GB:ABE84885.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:AAU44243.1); similar to Os01g0826900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001044677.1); contains InterPro domain Protein of unknown function DUF399; (InterPro:IPR007314)
PTAC4 (PLASTID TRANSCRIPTIONALLY ACTIVE4)
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G22890.2); similar to Os03g0857400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001051955.1); similar to Os08g0526300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001062297.1); similar to hypothetical protein OSJNBa0003M24.13 [Oryza sativa (japonica cultivar-group)] (GB:AAQ56398.1); contains domain no description (G3D.2.20.25.10); contains domain Zinc beta-ribbon (SSF57783)
NA
NA
NA
NA
JR700 (Arabidopsis thaliana receptor homology region transmembrane domain ring H2 motif protein 1); peptidase/ protein binding / zinc ion binding
serine carboxypeptidase S28 family protein
ATMINE1 (ARABIDOPSIS HOMOLOGUE OF BACTERIAL MINE 1); protein binding
NA
BPS1 (BYPASS 1)
similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD28388.1)
VAD1 (VASCULAR ASSOCIATED DEATH1)
glutaredoxin family protein
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1); similar to conserved hypothetical protein [Medicago truncatula] (GB:ABD32591.1)
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27030.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:AAN05517.1); similar to Os10g0463800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001064789.1)
calmodulin binding / lactoylglutathione lyase
similar to CW14 [Arabidopsis thaliana] (TAIR:AT1G59650.1); similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABF98316.1); contains InterPro domain Protein of unknown function DUF1336; (InterPro:IPR009769)
NA
NA
protein kinase
NA
VIP5 (VERNALIZATION INDEPENDENCE 5)
cinnamyl-alcohol dehydrogenase, putative
UBC16 (ubiquitin-conjugating enzyme 15); ubiquitin-protein ligase
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22230.1); similar to Os09g0370000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063016.1); similar to Os02g0753400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001048146.1); similar to hypothetical protein [Oryza sativa (japonica cultivar-group)] (GB:BAD15567.1)
NA
J8; heat shock protein binding / unfolded protein binding
XERICO; protein binding / zinc ion binding
NA
NA
NA
NA
NA
NA
NA
NA
NA
GCN5-related N-acetyltransferase (GNAT) family protein
amino acid transporter family protein
sigA-binding protein-related
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G07900.1); similar to Plant protein family, putative [Medicago truncatula] (GB:ABE79402.1); similar to H+-transporting two-sector ATPase, delta (OSCP) subunit; Hypothetical plant protein [Medicago truncatula] (GB:ABE83885.1); contains InterPro domain Protein of unknown function DUF246, plant; (InterPro:IPR004348)
NA
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G14390.1); similar to Esterase/lipase/thioesterase [Medicago truncatula] (GB:ABD32255.1); contains InterPro domain Esterase/lipase/thioesterase; (InterPro:IPR000379); contains InterPro domain Alpha/beta hydrolase fold-1; (InterPro:IPR000073)
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52720.1); similar to conserved hypothetical protein [Medicago truncatula] (GB:ABE92597.1)
phosphoric monoester hydrolase
S1 RNA-binding domain-containing protein
CCL (CCR-LIKE)
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41110.1); similar to Os09g0509400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063629.1); similar to Os08g0536100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001062361.1); similar to unnamed protein product; gene_id:MEE6.18 unknown protein-related [Medicago truncatula] (GB:ABE89396.1); contains domain FAMILY NOT NAMED (PTHR13199); contains domain SUBFAMILY NOT NAMED (PTHR13199:SF3)
NA
NA
late embryogenesis abundant protein, putative / LEA protein, putative
SAG21 (SENESCENCE-ASSOCIATED GENE 21)
NA
NA
similar to wound induced protein-like [Vitis vinifera] (GB:AAO12870.1)
NA
NA
IAGLU (INDOLE-3-ACETATE BETA-D-GLUCOSYLTRANSFERASE); UDP-glycosyltransferase/ transferase, transferring glycosyl groups
hydroxyproline-rich glycoprotein family protein
ACD1-LIKE; electron carrier
NA
similar to nucleoporin-related [Arabidopsis thaliana] (TAIR:AT5G20200.1); similar to conserved hypothetical protein [Medicago truncatula] (GB:ABE80407.1)
NA
NA
NA
(THIOREDOXIN-LIKE 5); thiol-disulfide exchange intermediate
D111/G-patch domain-containing protein
serine-rich protein-related
NA
NA
ATLCBK1 (A. THALIANA LONG-CHAIN BASE (LCB) KINASE 1); diacylglycerol kinase
NA
NA
glycerophosphoryl diester phosphodiesterase family protein
NA
NA
zinc finger (C3HC4-type RING finger) family protein
NA
NA
NA
MSL3 (MSCS-LIKE 3)
YGGT family protein
NA
mitochondrial substrate carrier family protein
NA
list of genes (text format)