GCRMA
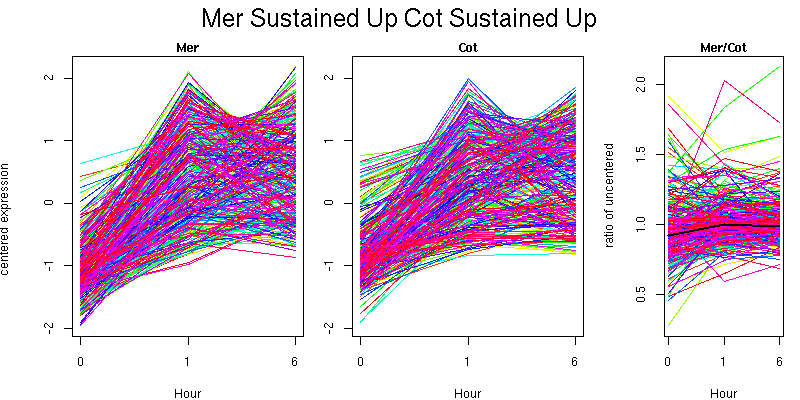
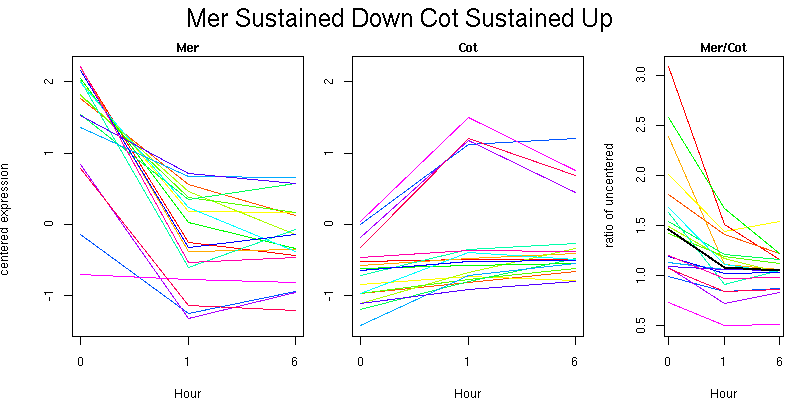
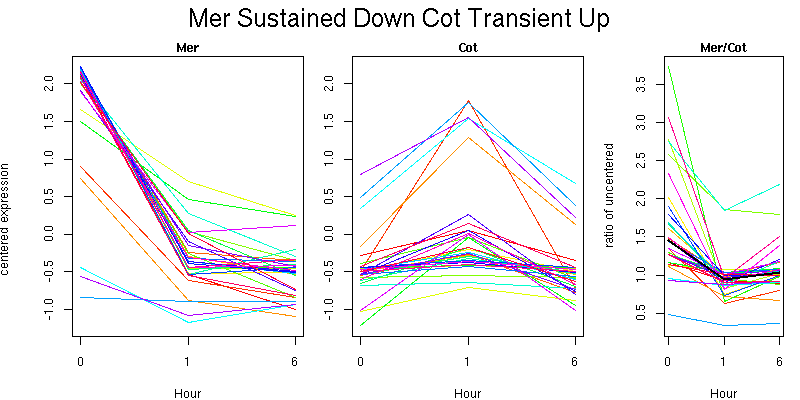
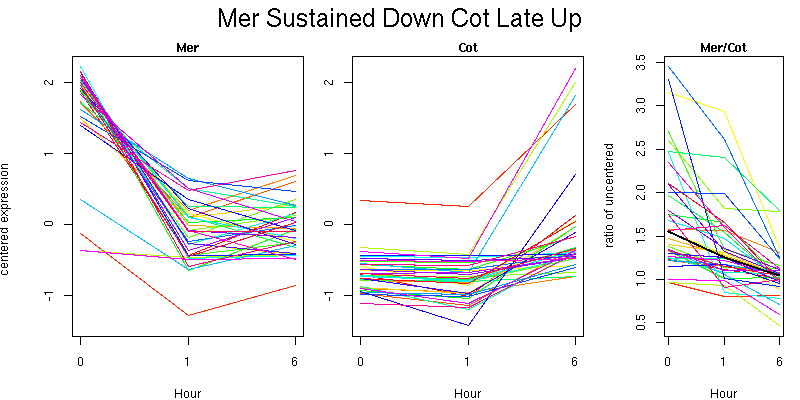
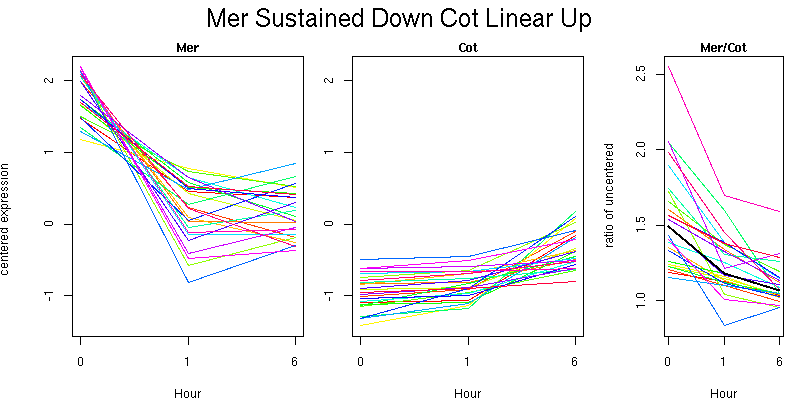
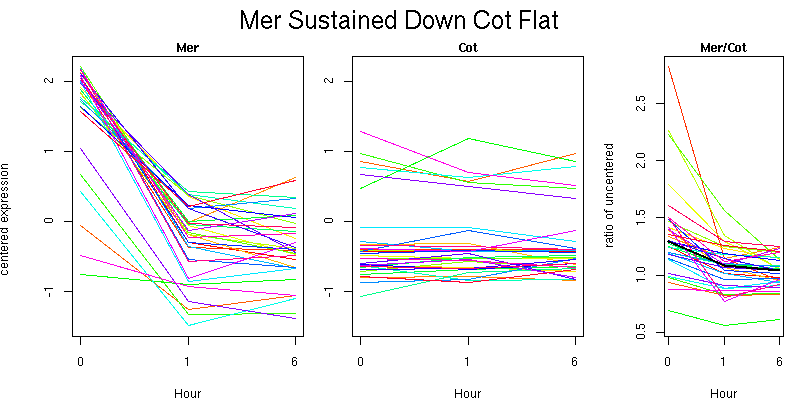
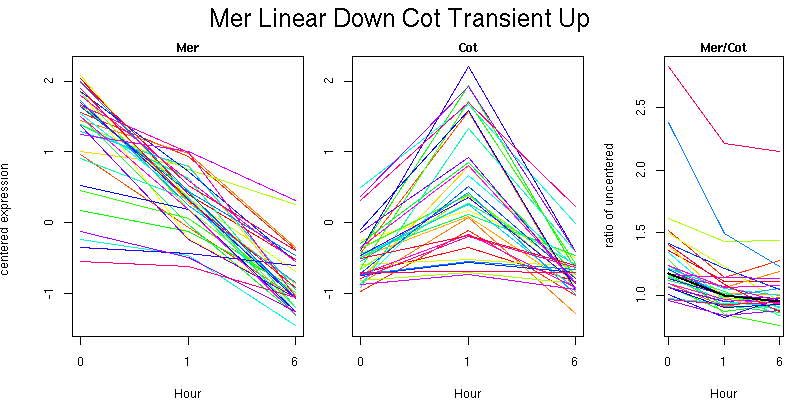
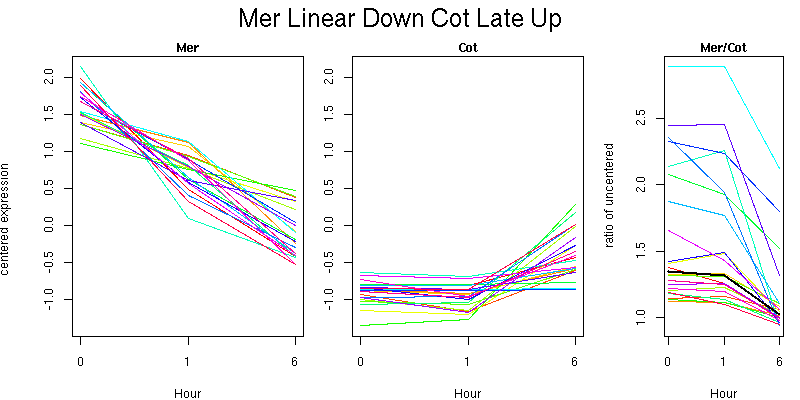
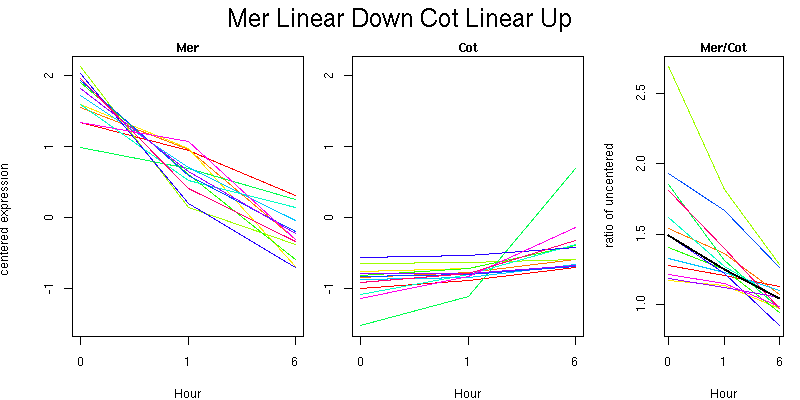
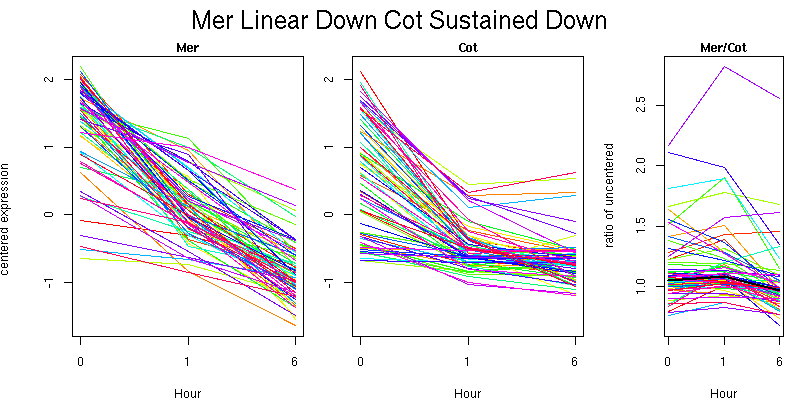
Pattern contains 333 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016877 | ligase activity, forming carbon-sulfur bonds | 4 | 0.35 | 0.00037 |
| GO:0016168 | chlorophyll binding | 4 | 0.37 | 0.00044 |
| GO:0016491 | oxidoreductase activity | 31 | 18.74 | 0.00415 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009628 | response to abiotic stimulus | 46 | 21.40 | 6.6e-07 |
| GO:0019748 | secondary metabolism | 17 | 4.80 | 7.3e-06 |
| GO:0009416 | response to light stimulus | 15 | 3.94 | 1.1e-05 |
| GO:0009314 | response to radiation | 15 | 3.99 | 1.2e-05 |
| GO:0006950 | response to stress | 35 | 17.46 | 7.0e-05 |
| GO:0009408 | response to heat | 7 | 1.07 | 9.4e-05 |
| GO:0050896 | response to stimulus | 53 | 31.46 | 1.0e-04 |
| GO:0009861 | jasmonic acid and ethylene-dependent systemic resistance | 9 | 1.95 | 1.5e-04 |
| GO:0009814 | defense response to pathogen, incompatible interaction | 11 | 2.97 | 2.0e-04 |
| GO:0007167 | enzyme linked receptor protein signaling pathway | 9 | 2.07 | 2.4e-04 |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 9 | 2.07 | 2.4e-04 |
| GO:0019438 | aromatic compound biosynthesis | 9 | 2.11 | 2.7e-04 |
| GO:0042221 | response to chemical stimulus | 29 | 14.51 | 3.2e-04 |
| GO:0010224 | response to UV-B | 4 | 0.34 | 3.3e-04 |
| GO:0007166 | cell surface receptor linked signal transduction | 10 | 2.66 | 3.5e-04 |
| GO:0009753 | response to jasmonic acid stimulus | 8 | 1.73 | 3.5e-04 |
| GO:0009698 | phenylpropanoid metabolism | 8 | 1.77 | 4.0e-04 |
| GO:0009611 | response to wounding | 10 | 2.71 | 4.1e-04 |
| GO:0009765 | photosynthesis, light harvesting | 4 | 0.37 | 4.7e-04 |
| GO:0009605 | response to external stimulus | 12 | 3.92 | 6.1e-04 |
| GO:0009411 | response to UV | 5 | 0.70 | 6.3e-04 |
| GO:0006725 | aromatic compound metabolism | 12 | 3.99 | 7.1e-04 |
| GO:0006979 | response to oxidative stress | 9 | 2.46 | 8.5e-04 |
| GO:0006800 | oxygen and reactive oxygen species metabolism | 9 | 2.69 | 1.6e-03 |
| GO:0046148 | pigment biosynthesis | 5 | 0.89 | 1.9e-03 |
| GO:0006720 | isoprenoid metabolism | 7 | 1.78 | 2.1e-03 |
| GO:0009699 | phenylpropanoid biosynthesis | 6 | 1.36 | 2.4e-03 |
| GO:0006575 | amino acid derivative metabolism | 9 | 2.87 | 2.4e-03 |
| GO:0006857 | oligopeptide transport | 5 | 0.97 | 2.9e-03 |
| GO:0015833 | peptide transport | 5 | 0.99 | 3.1e-03 |
| GO:0009058 | biosynthesis | 46 | 30.54 | 3.1e-03 |
| GO:0042440 | pigment metabolism | 5 | 1.04 | 3.8e-03 |
| GO:0007582 | physiological process | 178 | 155.23 | 4.3e-03 |
| GO:0015979 | photosynthesis | 6 | 1.57 | 5.0e-03 |
| GO:0009719 | response to endogenous stimulus | 21 | 11.31 | 5.0e-03 |
| GO:0009751 | response to salicylic acid stimulus | 6 | 1.62 | 5.8e-03 |
| GO:0044271 | nitrogen compound biosynthesis | 8 | 2.69 | 5.8e-03 |
| GO:0009110 | vitamin biosynthesis | 4 | 0.75 | 6.5e-03 |
| GO:0042398 | amino acid derivative biosynthesis | 7 | 2.19 | 6.5e-03 |
| GO:0009266 | response to temperature stimulus | 9 | 3.37 | 7.0e-03 |
| GO:0008610 | lipid biosynthesis | 10 | 4.02 | 7.4e-03 |
| GO:0042829 | defense response to pathogen | 12 | 5.33 | 7.7e-03 |
| GO:0044255 | cellular lipid metabolism | 12 | 5.38 | 8.2e-03 |
| GO:0006766 | vitamin metabolism | 5 | 1.28 | 9.2e-03 |
|
list of genes (text format)
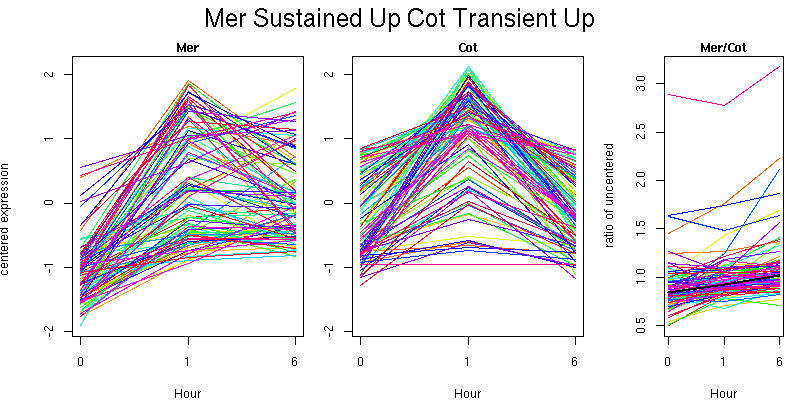
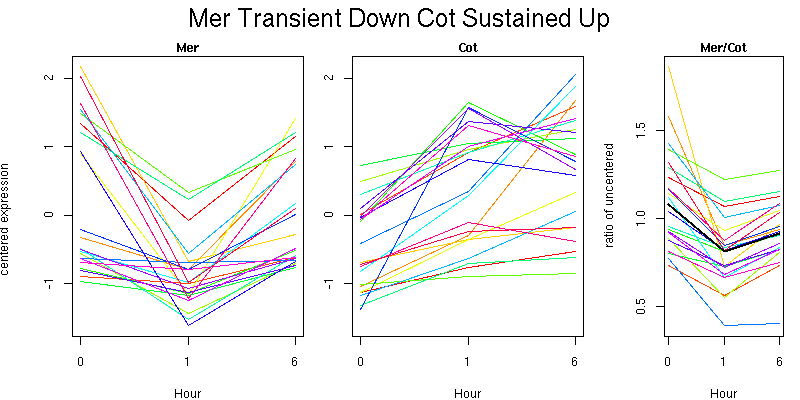
Pattern contains 104 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0015036 | disulfide oxidoreductase activity | 4 | 0.6 | 0.0031 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0019748 | secondary metabolism | 6 | 1.45 | 0.0033 |
| GO:0046483 | heterocycle metabolism | 4 | 0.72 | 0.0059 |
|
list of genes (text format)
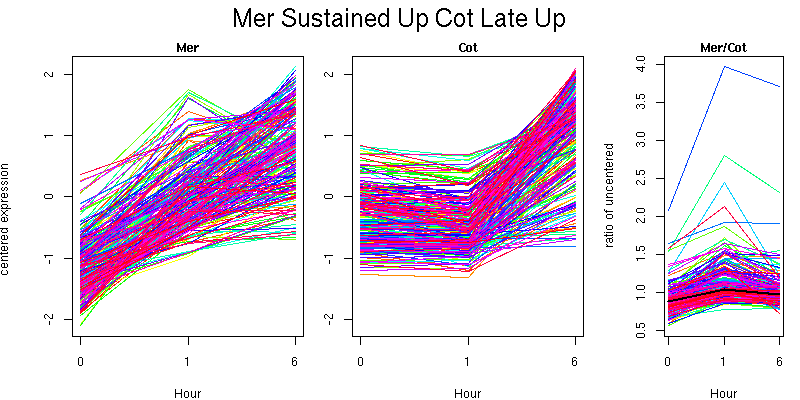
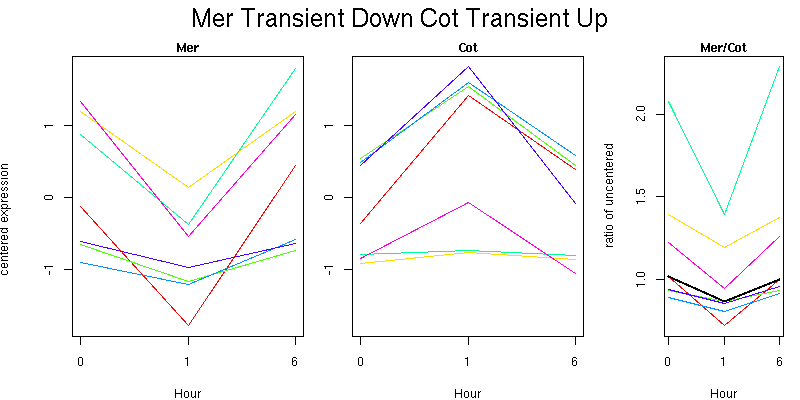
Pattern contains 270 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003735 | structural constituent of ribosome | 53 | 3.91 | 9.5e-45 |
| GO:0005198 | structural molecule activity | 54 | 5.25 | 5.0e-39 |
| GO:0030554 | adenyl nucleotide binding | 33 | 15.91 | 5.6e-05 |
| GO:0051082 | unfolded protein binding | 7 | 0.98 | 5.7e-05 |
| GO:0000166 | nucleotide binding | 39 | 20.76 | 9.6e-05 |
| GO:0017076 | purine nucleotide binding | 35 | 18.30 | 1.6e-04 |
| GO:0016538 | cyclin-dependent protein kinase regulator activity | 4 | 0.29 | 1.8e-04 |
| GO:0005524 | ATP binding | 31 | 15.56 | 1.9e-04 |
| GO:0019887 | protein kinase regulator activity | 4 | 0.45 | 9.9e-04 |
| GO:0019207 | kinase regulator activity | 4 | 0.46 | 1.1e-03 |
| GO:0008168 | methyltransferase activity | 7 | 1.65 | 1.4e-03 |
| GO:0016741 | transferase activity, transferring one-carbon groups | 7 | 1.68 | 1.5e-03 |
| GO:0016836 | hydro-lyase activity | 5 | 0.88 | 1.9e-03 |
| GO:0016874 | ligase activity | 11 | 3.93 | 2.1e-03 |
| GO:0008565 | protein transporter activity | 6 | 1.44 | 3.3e-03 |
| GO:0003899 | DNA-directed RNA polymerase activity | 4 | 0.64 | 3.9e-03 |
| GO:0003777 | microtubule motor activity | 4 | 0.84 | 1.0e-02 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0044249 | cellular biosynthesis | 83 | 22.80 | 8.2e-27 |
| GO:0009058 | biosynthesis | 85 | 25.05 | 2.1e-25 |
| GO:0006412 | protein biosynthesis | 58 | 11.41 | 2.2e-25 |
| GO:0006996 | organelle organization and biogenesis | 45 | 6.71 | 1.7e-24 |
| GO:0009059 | macromolecule biosynthesis | 62 | 14.17 | 1.1e-23 |
| GO:0042254 | ribosome biogenesis and assembly | 23 | 1.74 | 1.7e-19 |
| GO:0007028 | cytoplasm organization and biogenesis | 23 | 1.74 | 1.7e-19 |
| GO:0043170 | macromolecule metabolism | 110 | 50.62 | 6.8e-18 |
| GO:0019538 | protein metabolism | 90 | 36.06 | 7.9e-18 |
| GO:0016043 | cell organization and biogenesis | 52 | 13.96 | 8.8e-17 |
| GO:0007046 | ribosome biogenesis | 20 | 1.58 | 1.0e-16 |
| GO:0050875 | cellular physiological process | 173 | 114.70 | 1.8e-14 |
| GO:0044267 | cellular protein metabolism | 81 | 34.77 | 4.0e-14 |
| GO:0044238 | primary metabolism | 137 | 80.63 | 6.3e-14 |
| GO:0044260 | cellular macromolecule metabolism | 82 | 35.90 | 7.7e-14 |
| GO:0044237 | cellular metabolism | 145 | 88.46 | 9.7e-14 |
| GO:0009987 | cellular process | 173 | 118.70 | 8.1e-13 |
| GO:0007582 | physiological process | 178 | 127.34 | 1.5e-11 |
| GO:0008152 | metabolism | 150 | 100.10 | 6.3e-11 |
| GO:0006323 | DNA packaging | 12 | 1.54 | 4.7e-08 |
| GO:0006325 | establishment and/or maintenance of chromatin architecture | 12 | 1.54 | 4.7e-08 |
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 12 | 1.64 | 9.1e-08 |
| GO:0006520 | amino acid metabolism | 17 | 3.59 | 1.3e-07 |
| GO:0009308 | amine metabolism | 18 | 4.16 | 2.2e-07 |
| GO:0051276 | chromosome organization and biogenesis | 12 | 1.78 | 2.3e-07 |
| GO:0006259 | DNA metabolism | 17 | 4.10 | 8.5e-07 |
| GO:0006333 | chromatin assembly or disassembly | 9 | 1.08 | 1.3e-06 |
| GO:0006807 | nitrogen compound metabolism | 18 | 4.72 | 1.4e-06 |
| GO:0019752 | carboxylic acid metabolism | 22 | 6.82 | 1.5e-06 |
| GO:0006082 | organic acid metabolism | 22 | 6.85 | 1.6e-06 |
| GO:0031497 | chromatin assembly | 8 | 0.89 | 2.9e-06 |
| GO:0006519 | amino acid and derivative metabolism | 18 | 5.28 | 6.5e-06 |
| GO:0006334 | nucleosome assembly | 7 | 0.74 | 9.0e-06 |
| GO:0006414 | translational elongation | 5 | 0.32 | 1.4e-05 |
| GO:0006461 | protein complex assembly | 8 | 1.14 | 1.9e-05 |
| GO:0043037 | translation | 10 | 1.93 | 2.5e-05 |
| GO:0009309 | amine biosynthesis | 10 | 2.03 | 3.9e-05 |
| GO:0044271 | nitrogen compound biosynthesis | 10 | 2.21 | 7.8e-05 |
| GO:0008652 | amino acid biosynthesis | 9 | 1.82 | 9.2e-05 |
| GO:0006457 | protein folding | 10 | 2.51 | 2.3e-04 |
| GO:0009067 | aspartate family amino acid biosynthesis | 4 | 0.32 | 2.6e-04 |
| GO:0009081 | branched chain family amino acid metabolism | 4 | 0.32 | 2.6e-04 |
| GO:0009066 | aspartate family amino acid metabolism | 5 | 0.66 | 5.2e-04 |
| GO:0006261 | DNA-dependent DNA replication | 4 | 0.53 | 1.9e-03 |
| GO:0016568 | chromatin modification | 4 | 0.60 | 3.0e-03 |
| GO:0006260 | DNA replication | 5 | 1.08 | 4.5e-03 |
| GO:0007049 | cell cycle | 6 | 1.57 | 5.0e-03 |
|
list of genes (text format)
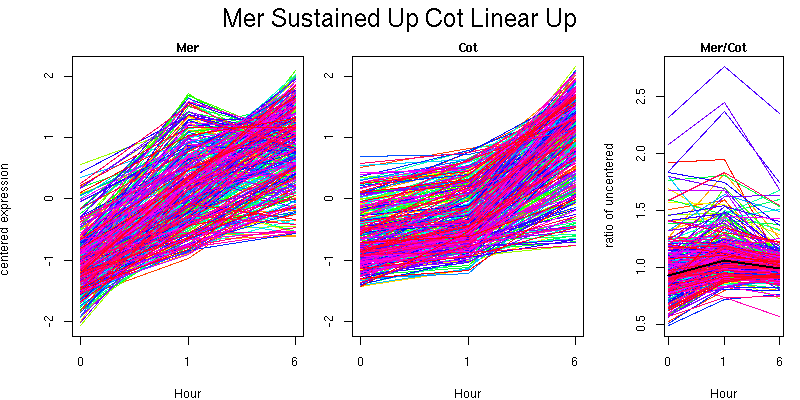
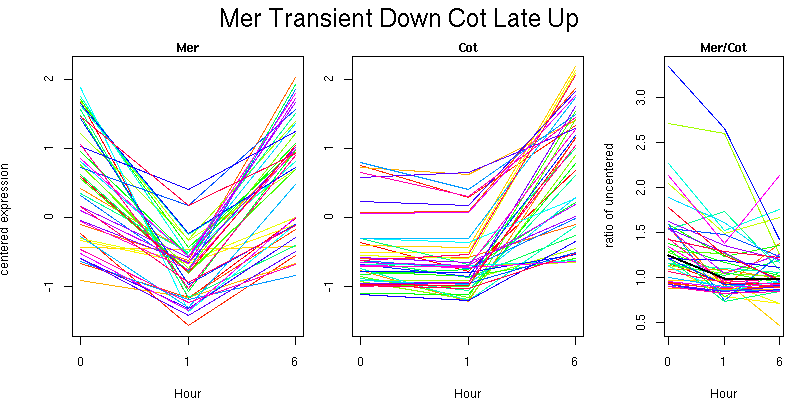
Pattern contains 362 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003735 | structural constituent of ribosome | 23 | 5.1 | 1.8e-09 |
| GO:0005198 | structural molecule activity | 26 | 6.8 | 6.0e-09 |
| GO:0003824 | catalytic activity | 131 | 110.6 | 9.8e-03 |
|
list of genes (text format)
Pattern contains 60 genes
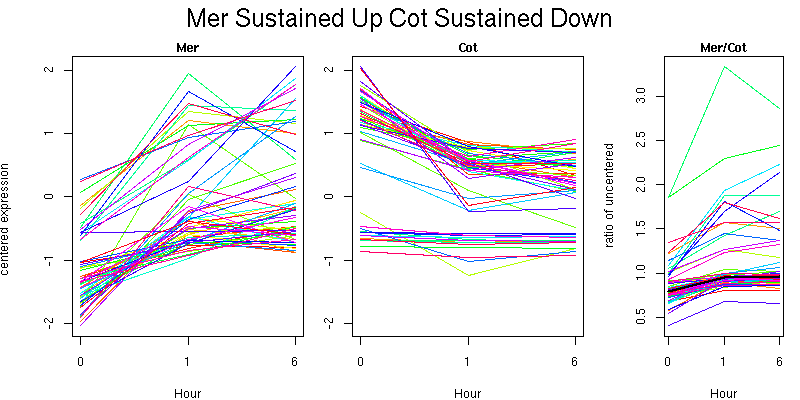
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005198 | structural molecule activity | 8 | 1.08 | 1.1e-05 |
| GO:0003735 | structural constituent of ribosome | 7 | 0.80 | 1.4e-05 |
| GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 4 | 0.25 | 1.1e-04 |
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | 4 | 0.30 | 2.2e-04 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006412 | protein biosynthesis | 9 | 2.36 | 0.00048 |
| GO:0044249 | cellular biosynthesis | 12 | 4.71 | 0.00193 |
| GO:0009059 | macromolecule biosynthesis | 9 | 2.93 | 0.00223 |
| GO:0019752 | carboxylic acid metabolism | 6 | 1.41 | 0.00270 |
| GO:0006082 | organic acid metabolism | 6 | 1.41 | 0.00276 |
| GO:0009058 | biosynthesis | 12 | 5.18 | 0.00422 |
| GO:0006520 | amino acid metabolism | 4 | 0.74 | 0.00639 |
|
list of genes (text format)
Pattern contains 46 genes
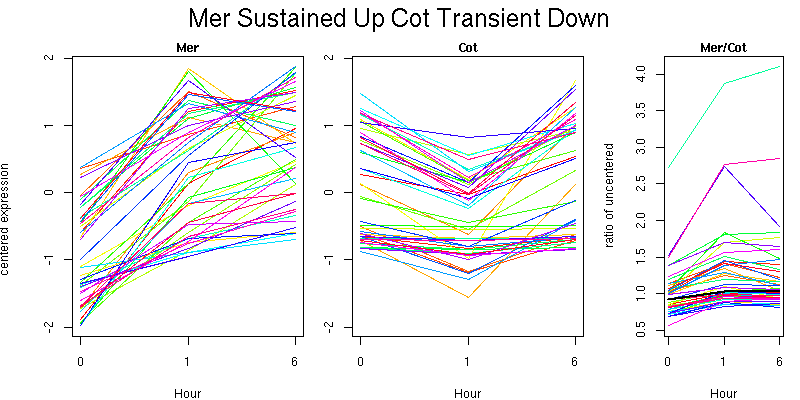
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003735 | structural constituent of ribosome | 5 | 0.66 | 0.00049 |
| GO:0005198 | structural molecule activity | 5 | 0.89 | 0.00182 |
|
Biological Pathway - no significant GO terms found
list of genes (text format)
Pattern contains 87 genes
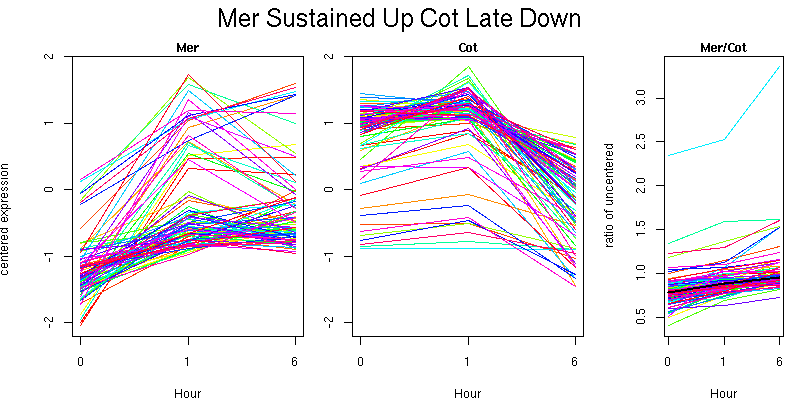
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 44 | 28.1 | 0.00023 |
| GO:0003723 | RNA binding | 6 | 1.7 | 0.00695 |
| GO:0016874 | ligase activity | 5 | 1.3 | 0.00958 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006399 | tRNA metabolism | 5 | 0.32 | 1.8e-05 |
| GO:0006418 | tRNA aminoacylation for protein translation | 4 | 0.20 | 4.4e-05 |
| GO:0043038 | amino acid activation | 4 | 0.20 | 4.4e-05 |
| GO:0043039 | tRNA aminoacylation | 4 | 0.20 | 4.4e-05 |
| GO:0006520 | amino acid metabolism | 7 | 1.08 | 1.0e-04 |
| GO:0016070 | RNA metabolism | 7 | 1.15 | 1.5e-04 |
| GO:0006519 | amino acid and derivative metabolism | 8 | 1.59 | 1.9e-04 |
| GO:0019752 | carboxylic acid metabolism | 9 | 2.06 | 2.0e-04 |
| GO:0006082 | organic acid metabolism | 9 | 2.07 | 2.1e-04 |
| GO:0009308 | amine metabolism | 7 | 1.26 | 2.6e-04 |
| GO:0043037 | translation | 5 | 0.58 | 2.9e-04 |
| GO:0006807 | nitrogen compound metabolism | 7 | 1.42 | 5.5e-04 |
| GO:0009793 | embryonic development (sensu Magnoliophyta) | 5 | 0.94 | 2.5e-03 |
| GO:0009790 | embryonic development | 5 | 1.00 | 3.3e-03 |
| GO:0048316 | seed development | 5 | 1.01 | 3.4e-03 |
| GO:0048608 | reproductive structure development | 5 | 1.02 | 3.6e-03 |
| GO:0008152 | metabolism | 42 | 30.19 | 3.8e-03 |
|
list of genes (text format)
Pattern contains 67 genes
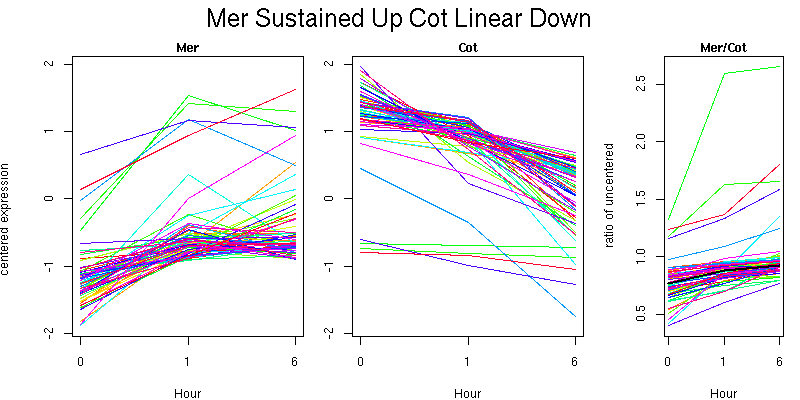
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003735 | structural constituent of ribosome | 9 | 0.91 | 2.8e-07 |
| GO:0005198 | structural molecule activity | 9 | 1.22 | 3.2e-06 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0044249 | cellular biosynthesis | 16 | 5.56 | 7.8e-05 |
| GO:0009058 | biosynthesis | 16 | 6.11 | 2.4e-04 |
| GO:0051188 | cofactor biosynthesis | 4 | 0.37 | 4.9e-04 |
| GO:0042254 | ribosome biogenesis and assembly | 4 | 0.42 | 8.6e-04 |
| GO:0007028 | cytoplasm organization and biogenesis | 4 | 0.42 | 8.6e-04 |
| GO:0006412 | protein biosynthesis | 9 | 2.78 | 1.7e-03 |
| GO:0019748 | secondary metabolism | 5 | 0.96 | 2.7e-03 |
| GO:0051186 | cofactor metabolism | 4 | 0.68 | 4.9e-03 |
| GO:0006807 | nitrogen compound metabolism | 5 | 1.15 | 5.8e-03 |
| GO:0009059 | macromolecule biosynthesis | 9 | 3.46 | 7.0e-03 |
|
list of genes (text format)
Pattern contains 89 genes
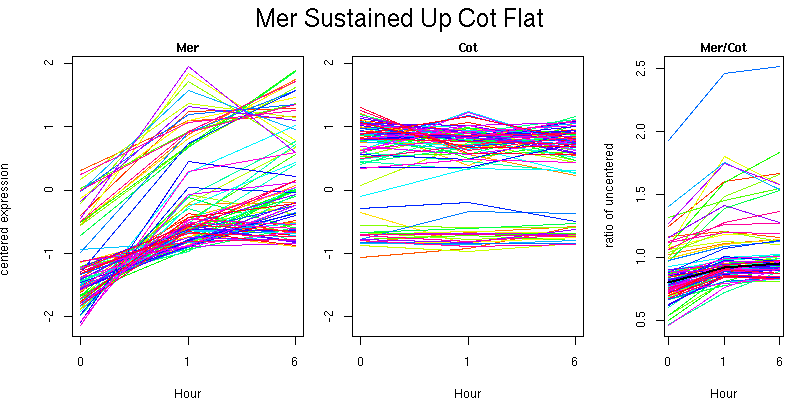
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016853 | isomerase activity | 5 | 0.73 | 0.00083 |
| GO:0009055 | electron carrier activity | 5 | 0.97 | 0.00288 |
|
list of genes (text format)
Pattern contains 64 genes
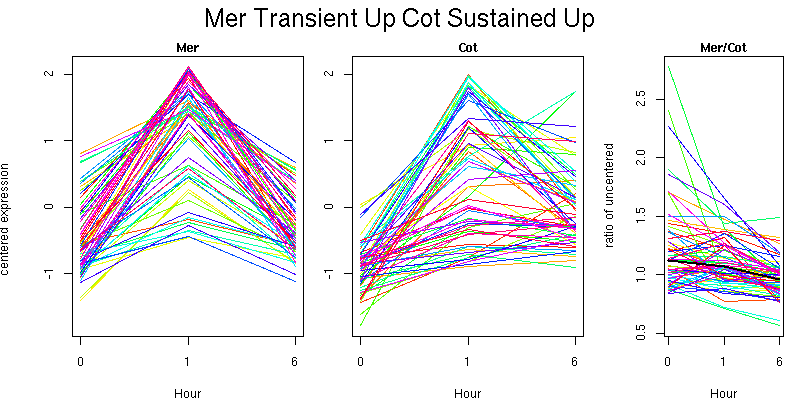
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0008509 | anion transporter activity | 4 | 0.27 | 0.00015 |
| GO:0003700 | transcription factor activity | 11 | 4.38 | 0.00375 |
| GO:0005215 | transporter activity | 9 | 3.40 | 0.00629 |
| GO:0015075 | ion transporter activity | 5 | 1.23 | 0.00760 |
| GO:0030528 | transcription regulator activity | 11 | 4.83 | 0.00780 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009628 | response to abiotic stimulus | 13 | 3.70 | 5.1e-05 |
| GO:0010035 | response to inorganic substance | 4 | 0.22 | 6.4e-05 |
| GO:0050896 | response to stimulus | 15 | 5.44 | 1.9e-04 |
| GO:0009651 | response to salt stress | 4 | 0.44 | 9.9e-04 |
| GO:0006970 | response to osmotic stress | 4 | 0.49 | 1.5e-03 |
| GO:0009416 | response to light stimulus | 4 | 0.68 | 4.7e-03 |
| GO:0009314 | response to radiation | 4 | 0.69 | 4.9e-03 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 104 genes
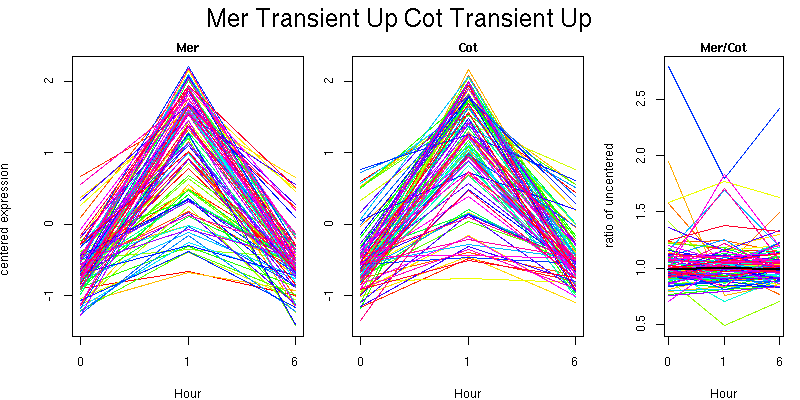
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003700 | transcription factor activity | 24 | 7.23 | 1.3e-07 |
| GO:0030528 | transcription regulator activity | 24 | 7.98 | 7.7e-07 |
| GO:0003677 | DNA binding | 24 | 9.45 | 1.5e-05 |
| GO:0003676 | nucleic acid binding | 27 | 13.20 | 1.7e-04 |
| GO:0016563 | transcriptional activator activity | 4 | 0.42 | 8.5e-04 |
| GO:0008270 | zinc ion binding | 12 | 4.28 | 1.2e-03 |
| GO:0005488 | binding | 46 | 32.56 | 3.3e-03 |
| GO:0046914 | transition metal ion binding | 14 | 6.19 | 3.4e-03 |
| GO:0015036 | disulfide oxidoreductase activity | 4 | 0.66 | 4.4e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0050789 | regulation of biological process | 21 | 7.80 | 2.0e-05 |
| GO:0050791 | regulation of physiological process | 20 | 7.28 | 2.5e-05 |
| GO:0009072 | aromatic amino acid family metabolism | 4 | 0.21 | 5.2e-05 |
| GO:0044237 | cellular metabolism | 49 | 31.07 | 5.9e-05 |
| GO:0042221 | response to chemical stimulus | 14 | 4.18 | 6.4e-05 |
| GO:0019222 | regulation of metabolism | 18 | 6.54 | 6.7e-05 |
| GO:0009628 | response to abiotic stimulus | 17 | 6.16 | 1.1e-04 |
| GO:0045449 | regulation of transcription | 17 | 6.25 | 1.3e-04 |
| GO:0019219 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 17 | 6.30 | 1.4e-04 |
| GO:0050896 | response to stimulus | 21 | 9.07 | 1.8e-04 |
| GO:0051244 | regulation of cellular physiological process | 18 | 7.10 | 1.9e-04 |
| GO:0050875 | cellular physiological process | 57 | 40.29 | 1.9e-04 |
| GO:0031323 | regulation of cellular metabolism | 17 | 6.48 | 2.0e-04 |
| GO:0050794 | regulation of cellular process | 18 | 7.14 | 2.1e-04 |
| GO:0009814 | defense response to pathogen, incompatible interaction | 6 | 0.85 | 2.2e-04 |
| GO:0006350 | transcription | 17 | 6.57 | 2.3e-04 |
| GO:0007582 | physiological process | 61 | 44.73 | 2.4e-04 |
| GO:0009987 | cellular process | 58 | 41.69 | 2.6e-04 |
| GO:0009719 | response to endogenous stimulus | 11 | 3.26 | 3.9e-04 |
| GO:0008152 | metabolism | 51 | 35.16 | 4.0e-04 |
| GO:0006950 | response to stress | 14 | 5.03 | 4.4e-04 |
| GO:0048518 | positive regulation of biological process | 4 | 0.37 | 5.3e-04 |
| GO:0009309 | amine biosynthesis | 5 | 0.71 | 7.5e-04 |
| GO:0042829 | defense response to pathogen | 7 | 1.54 | 8.7e-04 |
| GO:0044271 | nitrogen compound biosynthesis | 5 | 0.78 | 1.1e-03 |
| GO:0009611 | response to wounding | 5 | 0.78 | 1.1e-03 |
| GO:0009753 | response to jasmonic acid stimulus | 4 | 0.50 | 1.6e-03 |
| GO:0051707 | response to other organism | 8 | 2.31 | 2.2e-03 |
| GO:0009861 | jasmonic acid and ethylene-dependent systemic resistance | 4 | 0.56 | 2.4e-03 |
| GO:0042828 | response to pathogen | 7 | 1.85 | 2.5e-03 |
| GO:0006355 | regulation of transcription, DNA-dependent | 10 | 3.64 | 3.4e-03 |
| GO:0009308 | amine metabolism | 6 | 1.46 | 3.5e-03 |
| GO:0008652 | amino acid biosynthesis | 4 | 0.64 | 3.9e-03 |
| GO:0009613 | response to pest, pathogen or parasite | 7 | 2.01 | 4.0e-03 |
| GO:0006351 | transcription, DNA-dependent | 10 | 3.76 | 4.2e-03 |
| GO:0009605 | response to external stimulus | 5 | 1.13 | 5.5e-03 |
| GO:0009416 | response to light stimulus | 5 | 1.13 | 5.6e-03 |
| GO:0044238 | primary metabolism | 40 | 28.32 | 5.8e-03 |
| GO:0006725 | aromatic compound metabolism | 5 | 1.15 | 5.9e-03 |
| GO:0009314 | response to radiation | 5 | 1.15 | 5.9e-03 |
| GO:0006807 | nitrogen compound metabolism | 6 | 1.66 | 6.3e-03 |
| GO:0009651 | response to salt stress | 4 | 0.74 | 6.4e-03 |
| GO:0006952 | defense response | 7 | 2.30 | 8.1e-03 |
| GO:0009607 | response to biotic stimulus | 8 | 2.90 | 8.4e-03 |
| GO:0006520 | amino acid metabolism | 5 | 1.26 | 8.7e-03 |
| GO:0006970 | response to osmotic stress | 4 | 0.82 | 9.3e-03 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 18 | 10.08 | 9.9e-03 |
|
list of genes (text format)
Pattern contains 9 genes
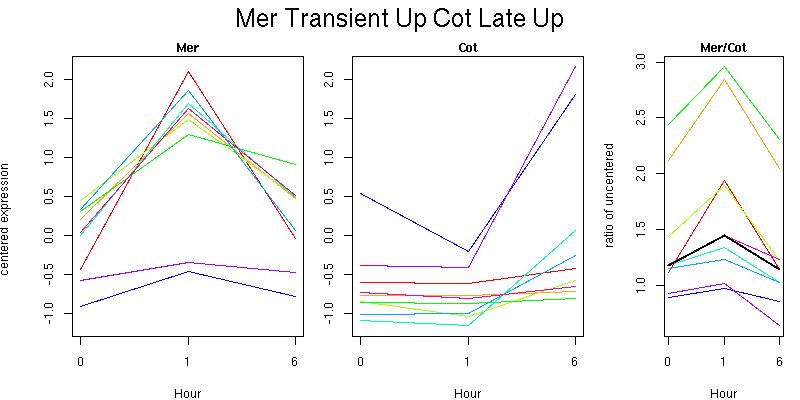
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016740 | transferase activity | 4 | 0.71 | 0.0025 |
|
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 19 genes
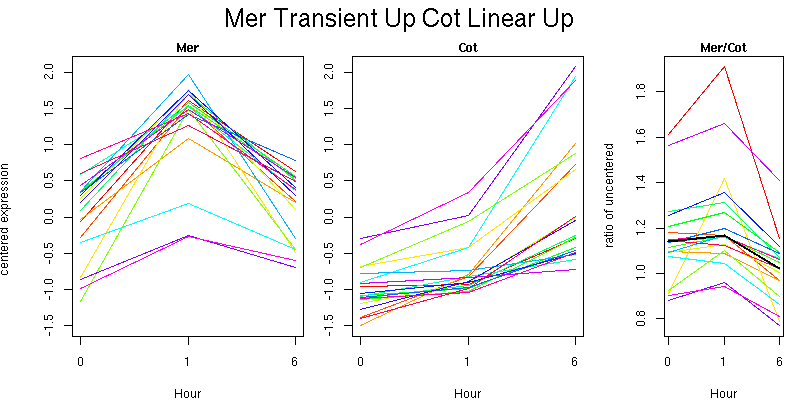
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0044262 | cellular carbohydrate metabolism | 4 | 0.38 | 0.00044 |
| GO:0009987 | cellular process | 15 | 8.34 | 0.00091 |
| GO:0007582 | physiological process | 15 | 8.95 | 0.00229 |
| GO:0005975 | carbohydrate metabolism | 4 | 0.62 | 0.00286 |
| GO:0050875 | cellular physiological process | 14 | 8.06 | 0.00342 |
| GO:0009058 | biosynthesis | 6 | 1.76 | 0.00552 |
|
list of genes (text format)
Pattern contains 12 genes
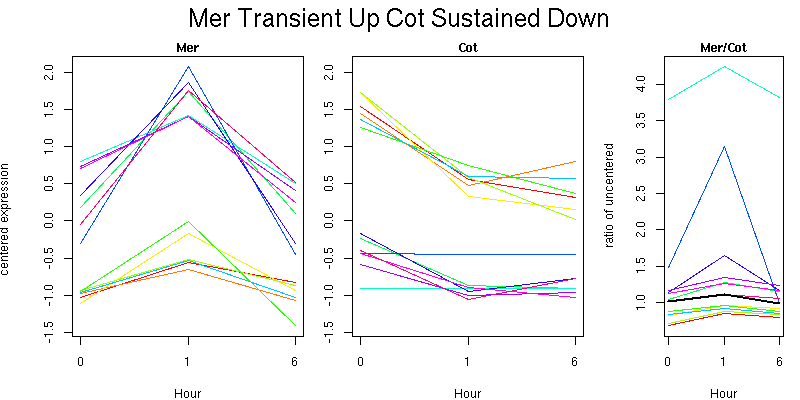
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 14 genes
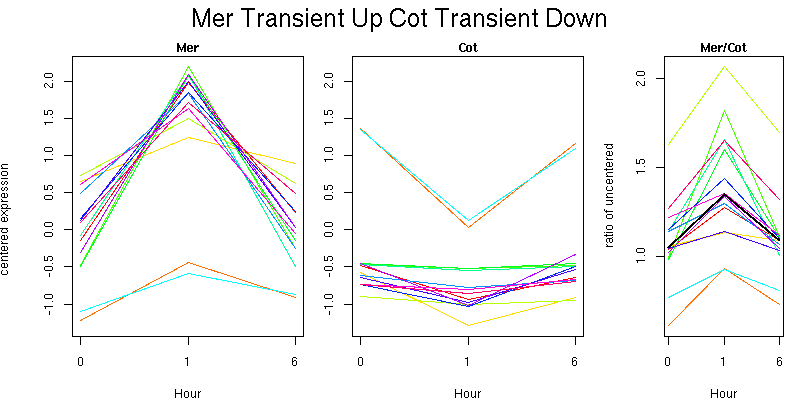
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 94 genes
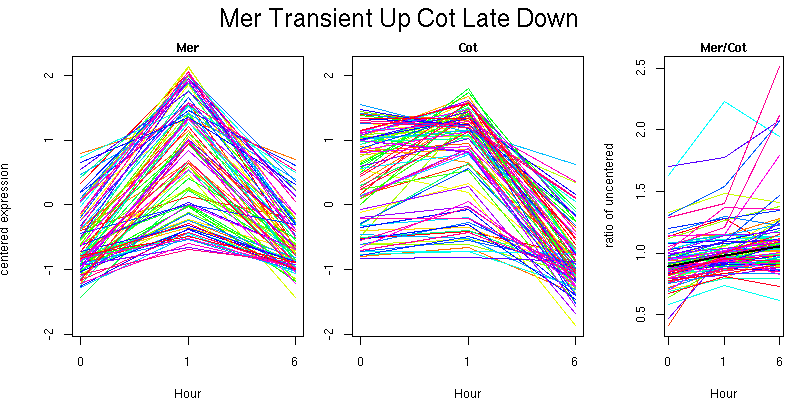
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 47 | 30 | 0.00024 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006629 | lipid metabolism | 10 | 2.17 | 5.9e-05 |
| GO:0006790 | sulfur metabolism | 4 | 0.33 | 3.3e-04 |
| GO:0019752 | carboxylic acid metabolism | 8 | 2.28 | 2.0e-03 |
| GO:0006082 | organic acid metabolism | 8 | 2.29 | 2.0e-03 |
| GO:0008152 | metabolism | 45 | 33.50 | 6.8e-03 |
| GO:0006519 | amino acid and derivative metabolism | 6 | 1.77 | 8.5e-03 |
|
list of genes (text format)
Pattern contains 71 genes
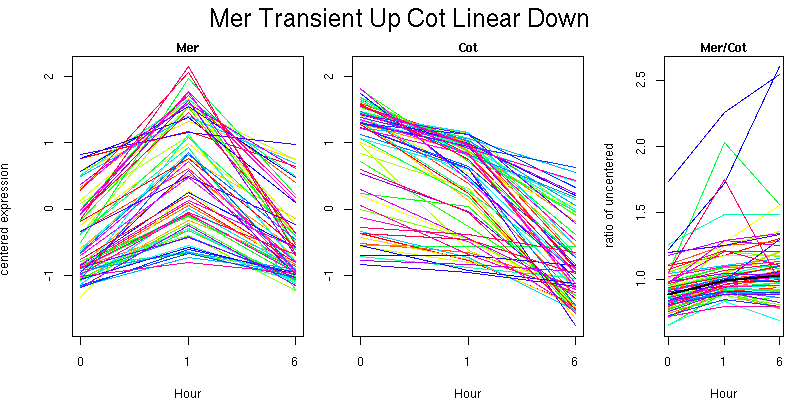
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 34 | 22 | 0.0014 |
| GO:0016491 | oxidoreductase activity | 10 | 4 | 0.0058 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0019752 | carboxylic acid metabolism | 9 | 1.86 | 9.3e-05 |
| GO:0006082 | organic acid metabolism | 9 | 1.87 | 9.6e-05 |
| GO:0006092 | main pathways of carbohydrate metabolism | 4 | 0.46 | 1.2e-03 |
| GO:0008152 | metabolism | 40 | 27.30 | 1.2e-03 |
| GO:0009793 | embryonic development (sensu Magnoliophyta) | 5 | 0.85 | 1.6e-03 |
| GO:0009790 | embryonic development | 5 | 0.91 | 2.1e-03 |
| GO:0044237 | cellular metabolism | 36 | 24.13 | 2.1e-03 |
| GO:0048316 | seed development | 5 | 0.91 | 2.2e-03 |
| GO:0006979 | response to oxidative stress | 4 | 0.55 | 2.3e-03 |
| GO:0048608 | reproductive structure development | 5 | 0.92 | 2.3e-03 |
| GO:0006520 | amino acid metabolism | 5 | 0.98 | 3.0e-03 |
| GO:0006800 | oxygen and reactive oxygen species metabolism | 4 | 0.60 | 3.1e-03 |
| GO:0044262 | cellular carbohydrate metabolism | 6 | 1.46 | 3.3e-03 |
| GO:0007582 | physiological process | 46 | 34.73 | 3.5e-03 |
| GO:0006457 | protein folding | 4 | 0.69 | 4.9e-03 |
| GO:0009308 | amine metabolism | 5 | 1.14 | 5.5e-03 |
| GO:0015980 | energy derivation by oxidation of organic compounds | 4 | 0.71 | 5.7e-03 |
| GO:0050875 | cellular physiological process | 42 | 31.28 | 5.7e-03 |
| GO:0000003 | reproduction | 5 | 1.20 | 7.0e-03 |
| GO:0006807 | nitrogen compound metabolism | 5 | 1.29 | 9.3e-03 |
|
list of genes (text format)
Pattern contains 21 genes
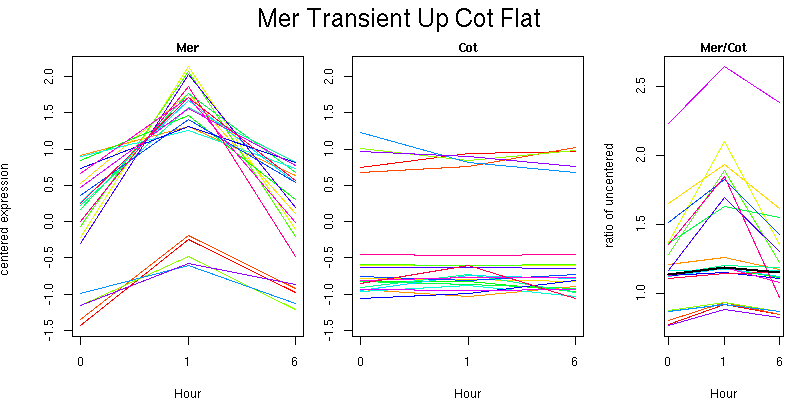
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 12 | 5.5 | 0.0010 |
| GO:0016740 | transferase activity | 6 | 1.9 | 0.0078 |
|
list of genes (text format)
Pattern contains 22 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 7 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 138 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 67 | 44.2 | 2.5e-05 |
| GO:0016491 | oxidoreductase activity | 17 | 8.0 | 2.6e-03 |
| GO:0020037 | heme binding | 5 | 1.3 | 8.7e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006790 | sulfur metabolism | 4 | 0.49 | 0.0015 |
| GO:0006807 | nitrogen compound metabolism | 8 | 2.36 | 0.0026 |
| GO:0006091 | generation of precursor metabolites and energy | 12 | 4.90 | 0.0037 |
| GO:0006519 | amino acid and derivative metabolism | 8 | 2.64 | 0.0051 |
| GO:0008152 | metabolism | 64 | 50.05 | 0.0067 |
| GO:0019752 | carboxylic acid metabolism | 9 | 3.41 | 0.0073 |
| GO:0006082 | organic acid metabolism | 9 | 3.42 | 0.0075 |
|
list of genes (text format)
Pattern contains 86 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016758 | transferase activity, transferring hexosyl groups | 5 | 0.83 | 0.0014 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0015980 | energy derivation by oxidation of organic compounds | 4 | 0.75 | 0.0067 |
| GO:0006066 | alcohol metabolism | 4 | 0.77 | 0.0073 |
|
list of genes (text format)
Pattern contains 8 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
list of genes (text format)
Pattern contains 26 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 6 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
list of genes (text format)
Pattern contains 15 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0044444 | cytoplasmic part | 9 | 4.3 | 0.0085 |
|
list of genes (text format)
Pattern contains 15 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
list of genes (text format)
Pattern contains 90 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0030508 | thiol-disulfide exchange intermediate activity | 4 | 0.27 | 0.00015 |
| GO:0015035 | protein disulfide oxidoreductase activity | 4 | 0.31 | 0.00025 |
| GO:0015036 | disulfide oxidoreductase activity | 4 | 0.48 | 0.00139 |
| GO:0009055 | electron carrier activity | 5 | 0.86 | 0.00172 |
| GO:0016491 | oxidoreductase activity | 11 | 4.47 | 0.00464 |
|
Biological Pathway - no significant GO terms found
list of genes (text format)
Pattern contains 16 genes
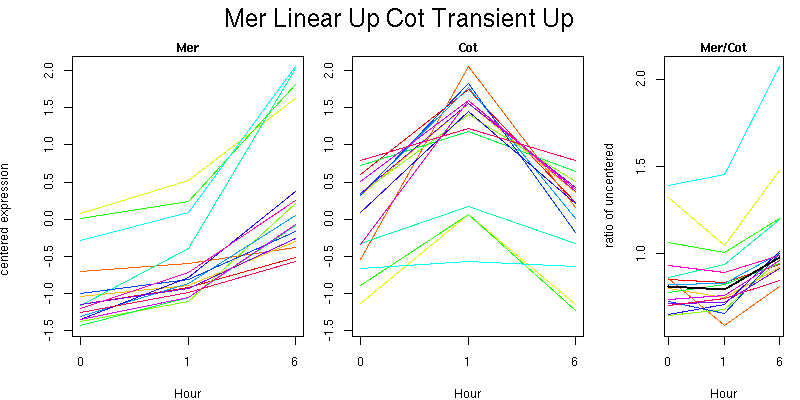
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 308 genes
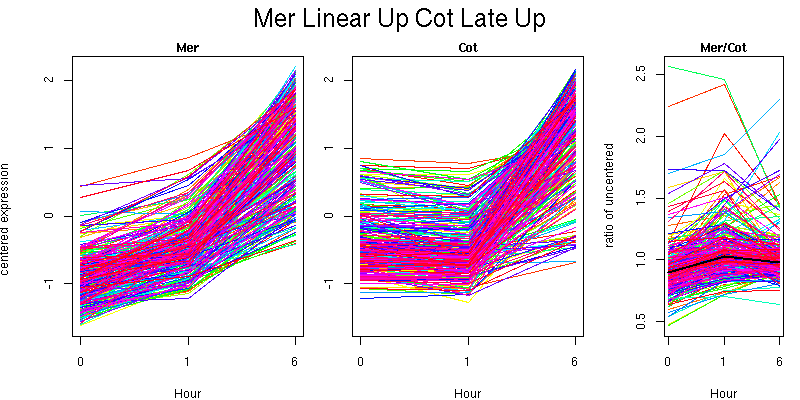
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005527 | macrolide binding | 5 | 0.32 | 1.4e-05 |
| GO:0005528 | FK506 binding | 5 | 0.32 | 1.4e-05 |
| GO:0008168 | methyltransferase activity | 10 | 1.85 | 1.7e-05 |
| GO:0016741 | transferase activity, transferring one-carbon groups | 10 | 1.88 | 1.9e-05 |
| GO:0008144 | drug binding | 5 | 0.35 | 2.2e-05 |
| GO:0008170 | N-methyltransferase activity | 4 | 0.26 | 1.2e-04 |
| GO:0003735 | structural constituent of ribosome | 13 | 4.37 | 4.8e-04 |
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | 5 | 0.76 | 9.7e-04 |
| GO:0016859 | cis-trans isomerase activity | 5 | 0.78 | 1.1e-03 |
| GO:0015450 | protein translocase activity | 4 | 0.60 | 3.0e-03 |
| GO:0008565 | protein transporter activity | 6 | 1.61 | 5.7e-03 |
| GO:0005198 | structural molecule activity | 13 | 5.86 | 6.4e-03 |
| GO:0005215 | transporter activity | 26 | 15.73 | 8.4e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006996 | organelle organization and biogenesis | 26 | 7.33 | 2.5e-08 |
| GO:0042254 | ribosome biogenesis and assembly | 11 | 1.90 | 3.3e-06 |
| GO:0007028 | cytoplasm organization and biogenesis | 11 | 1.90 | 3.3e-06 |
| GO:0016043 | cell organization and biogenesis | 35 | 15.23 | 3.7e-06 |
| GO:0007046 | ribosome biogenesis | 10 | 1.73 | 9.2e-06 |
| GO:0006334 | nucleosome assembly | 7 | 0.81 | 1.6e-05 |
| GO:0031497 | chromatin assembly | 7 | 0.97 | 5.2e-05 |
| GO:0051276 | chromosome organization and biogenesis | 9 | 1.94 | 1.5e-04 |
| GO:0009058 | biosynthesis | 47 | 27.33 | 1.5e-04 |
| GO:0006333 | chromatin assembly or disassembly | 7 | 1.17 | 1.7e-04 |
| GO:0009112 | nucleobase metabolism | 4 | 0.30 | 2.1e-04 |
| GO:0006461 | protein complex assembly | 7 | 1.25 | 2.5e-04 |
| GO:0006323 | DNA packaging | 8 | 1.68 | 2.9e-04 |
| GO:0006325 | establishment and/or maintenance of chromatin architecture | 8 | 1.68 | 2.9e-04 |
| GO:0009152 | purine ribonucleotide biosynthesis | 5 | 0.64 | 4.2e-04 |
| GO:0007001 | chromosome organization and biogenesis (sensu Eukaryota) | 8 | 1.78 | 4.3e-04 |
| GO:0009260 | ribonucleotide biosynthesis | 5 | 0.65 | 4.7e-04 |
| GO:0009150 | purine ribonucleotide metabolism | 5 | 0.67 | 5.2e-04 |
| GO:0009165 | nucleotide biosynthesis | 6 | 1.02 | 5.3e-04 |
| GO:0006259 | DNA metabolism | 13 | 4.47 | 5.9e-04 |
| GO:0006163 | purine nucleotide metabolism | 5 | 0.71 | 7.0e-04 |
| GO:0006164 | purine nucleotide biosynthesis | 5 | 0.71 | 7.0e-04 |
| GO:0044249 | cellular biosynthesis | 41 | 24.88 | 9.8e-04 |
| GO:0009259 | ribonucleotide metabolism | 5 | 0.78 | 1.1e-03 |
| GO:0006364 | rRNA processing | 4 | 0.51 | 1.6e-03 |
| GO:0016072 | rRNA metabolism | 4 | 0.54 | 2.0e-03 |
| GO:0042440 | pigment metabolism | 5 | 0.93 | 2.3e-03 |
| GO:0009987 | cellular process | 152 | 129.49 | 3.1e-03 |
| GO:0015031 | protein transport | 11 | 4.16 | 3.3e-03 |
| GO:0017038 | protein import | 4 | 0.64 | 3.7e-03 |
| GO:0045184 | establishment of protein localization | 11 | 4.31 | 4.2e-03 |
| GO:0008104 | protein localization | 11 | 4.31 | 4.2e-03 |
| GO:0009309 | amine biosynthesis | 7 | 2.22 | 7.0e-03 |
| GO:0050875 | cellular physiological process | 145 | 125.12 | 8.1e-03 |
| GO:0046148 | pigment biosynthesis | 4 | 0.80 | 8.3e-03 |
| GO:0009117 | nucleotide metabolism | 6 | 1.76 | 8.4e-03 |
| GO:0006605 | protein targeting | 5 | 1.26 | 8.7e-03 |
|
list of genes (text format)
Pattern contains 314 genes
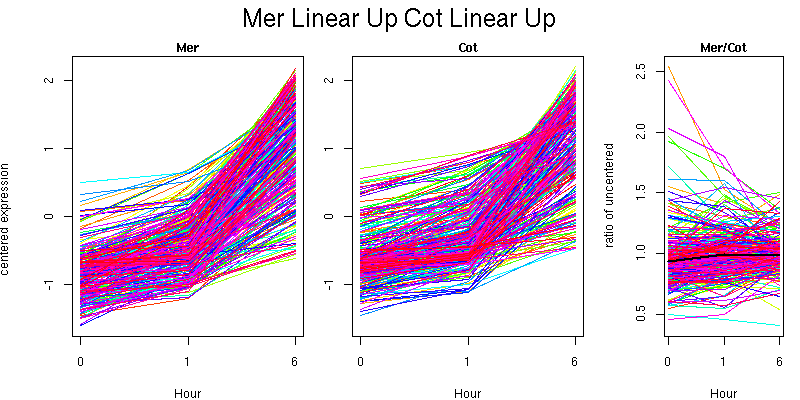
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0009055 | electron carrier activity | 13 | 3.4 | 4.6e-05 |
| GO:0005525 | GTP binding | 11 | 2.9 | 1.8e-04 |
| GO:0019001 | guanyl nucleotide binding | 11 | 3.0 | 2.1e-04 |
| GO:0030234 | enzyme regulator activity | 11 | 3.4 | 6.1e-04 |
| GO:0003824 | catalytic activity | 125 | 98.6 | 7.1e-04 |
| GO:0015035 | protein disulfide oxidoreductase activity | 6 | 1.2 | 1.5e-03 |
| GO:0015036 | disulfide oxidoreductase activity | 7 | 1.9 | 3.3e-03 |
| GO:0005507 | copper ion binding | 6 | 1.5 | 3.4e-03 |
| GO:0030508 | thiol-disulfide exchange intermediate activity | 5 | 1.1 | 4.5e-03 |
| GO:0016830 | carbon-carbon lyase activity | 5 | 1.2 | 7.0e-03 |
| GO:0016491 | oxidoreductase activity | 29 | 17.9 | 7.1e-03 |
| GO:0005351 | sugar porter activity | 5 | 1.2 | 8.2e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0015979 | photosynthesis | 11 | 1.46 | 2.2e-07 |
| GO:0019684 | photosynthesis, light reaction | 8 | 0.89 | 2.6e-06 |
| GO:0000074 | regulation of progression through cell cycle | 7 | 1.12 | 1.3e-04 |
| GO:0051726 | regulation of cell cycle | 7 | 1.12 | 1.3e-04 |
| GO:0007049 | cell cycle | 8 | 1.77 | 4.1e-04 |
| GO:0007582 | physiological process | 171 | 143.66 | 4.9e-04 |
| GO:0006091 | generation of precursor metabolites and energy | 22 | 11.05 | 1.8e-03 |
| GO:0006519 | amino acid and derivative metabolism | 14 | 5.96 | 2.8e-03 |
| GO:0044271 | nitrogen compound biosynthesis | 8 | 2.49 | 3.6e-03 |
| GO:0006810 | transport | 42 | 27.80 | 4.4e-03 |
| GO:0051179 | localization | 42 | 27.98 | 4.9e-03 |
| GO:0051234 | establishment of localization | 42 | 27.98 | 4.9e-03 |
| GO:0044249 | cellular biosynthesis | 39 | 25.73 | 5.7e-03 |
| GO:0009058 | biosynthesis | 42 | 28.26 | 5.8e-03 |
| GO:0006807 | nitrogen compound metabolism | 12 | 5.33 | 7.6e-03 |
| GO:0006869 | lipid transport | 5 | 1.26 | 8.6e-03 |
| GO:0050875 | cellular physiological process | 149 | 129.39 | 9.8e-03 |
|
list of genes (text format)
Pattern contains 22 genes
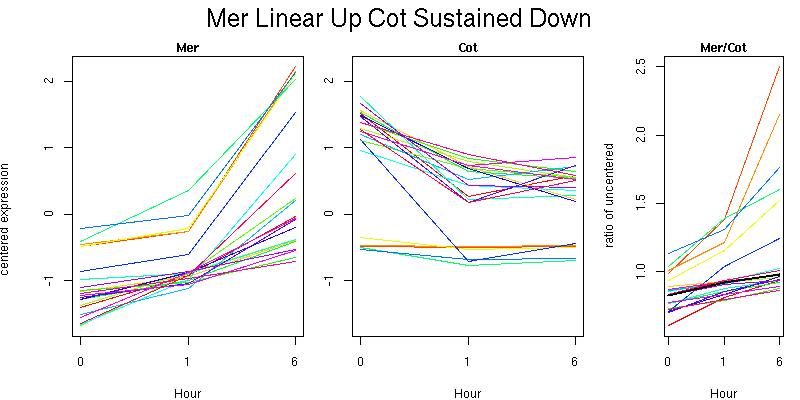
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
list of genes (text format)
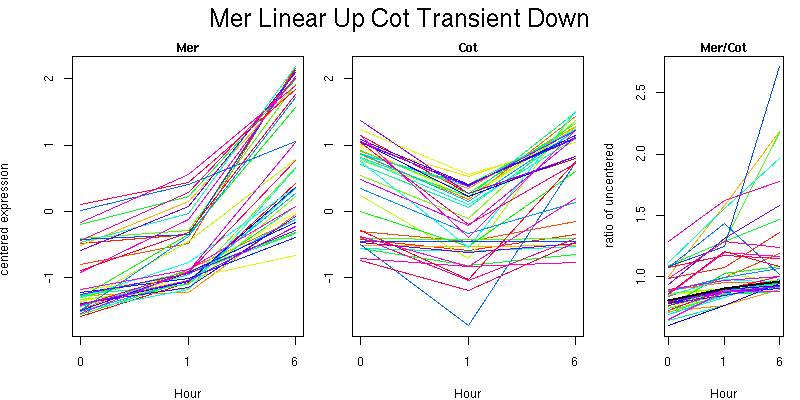
Pattern contains 42 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
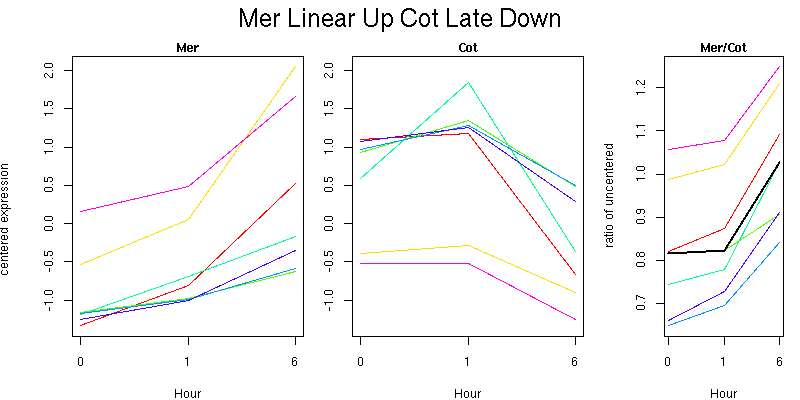
list of genes (text format)
Pattern contains 7 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
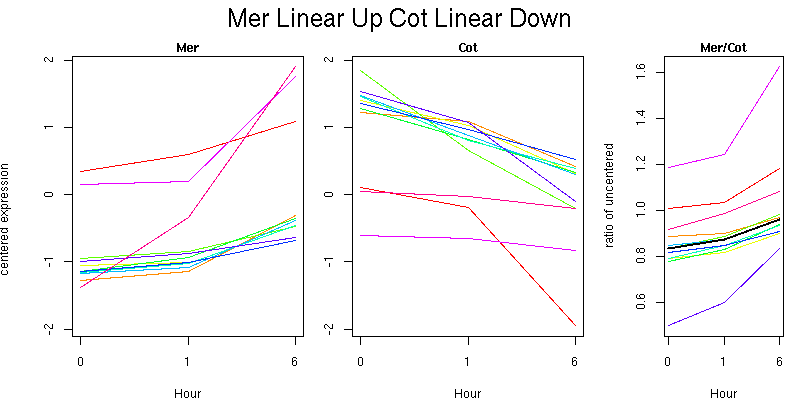
list of genes (text format)
Pattern contains 11 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
list of genes (text format)
Pattern contains 41 genes
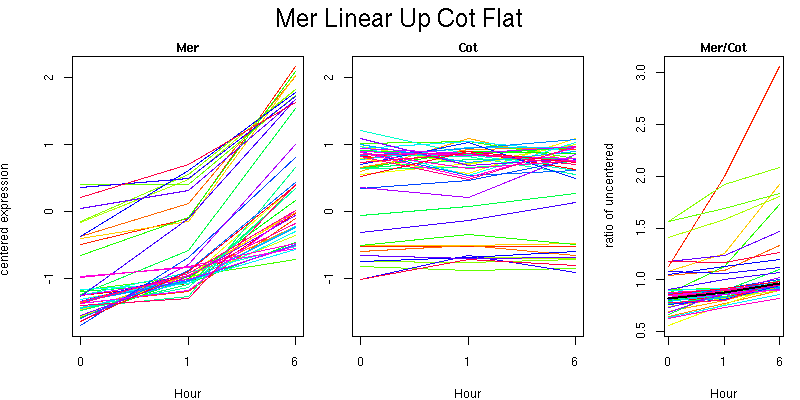
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0015036 | disulfide oxidoreductase activity | 4 | 0.24 | 9.6e-05 |
|
Biological Pathway - no significant GO terms found
list of genes (text format)
Pattern contains 18 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 31 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0007154 | cell communication | 5 | 1 | 0.0026 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 37 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009628 | response to abiotic stimulus | 8 | 2.4 | 0.0020 |
| GO:0042221 | response to chemical stimulus | 6 | 1.6 | 0.0049 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 25 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005215 | transporter activity | 5 | 1.2 | 0.0055 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009605 | response to external stimulus | 4 | 0.31 | 0.00022 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 303 genes
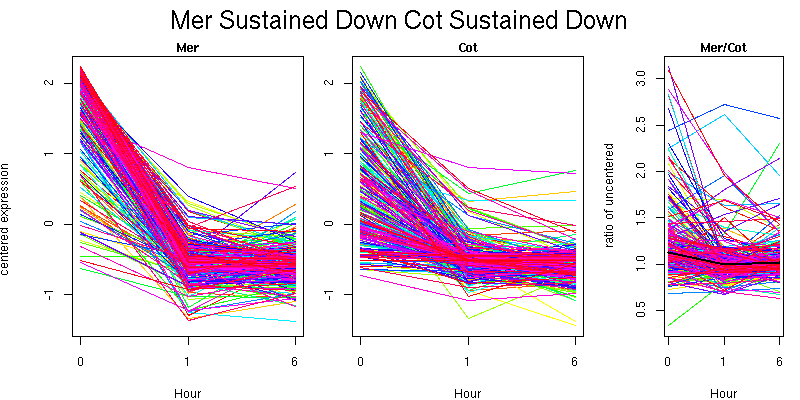
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0050791 | regulation of physiological process | 56 | 21.59 | 2.2e-11 |
| GO:0051244 | regulation of cellular physiological process | 55 | 21.06 | 2.7e-11 |
| GO:0050794 | regulation of cellular process | 55 | 21.17 | 3.4e-11 |
| GO:0019222 | regulation of metabolism | 52 | 19.38 | 4.1e-11 |
| GO:0031323 | regulation of cellular metabolism | 51 | 19.20 | 9.4e-11 |
| GO:0045449 | regulation of transcription | 49 | 18.54 | 2.8e-10 |
| GO:0050789 | regulation of biological process | 56 | 23.14 | 3.2e-10 |
| GO:0019219 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 49 | 18.69 | 3.7e-10 |
| GO:0006350 | transcription | 50 | 19.48 | 4.9e-10 |
| GO:0009725 | response to hormone stimulus | 24 | 6.81 | 9.9e-08 |
| GO:0009719 | response to endogenous stimulus | 27 | 9.66 | 1.5e-06 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 55 | 29.89 | 4.8e-06 |
| GO:0009733 | response to auxin stimulus | 13 | 2.81 | 5.3e-06 |
| GO:0006351 | transcription, DNA-dependent | 28 | 11.15 | 7.3e-06 |
| GO:0006355 | regulation of transcription, DNA-dependent | 27 | 10.79 | 1.2e-05 |
| GO:0042221 | response to chemical stimulus | 27 | 12.39 | 1.2e-04 |
| GO:0009628 | response to abiotic stimulus | 34 | 18.28 | 3.4e-04 |
| GO:0009735 | response to cytokinin stimulus | 5 | 0.72 | 7.5e-04 |
| GO:0006944 | membrane fusion | 4 | 0.44 | 9.5e-04 |
| GO:0009723 | response to ethylene stimulus | 7 | 1.61 | 1.2e-03 |
| GO:0009755 | hormone-mediated signaling | 8 | 2.13 | 1.4e-03 |
| GO:0050896 | response to stimulus | 41 | 26.88 | 4.0e-03 |
| GO:0016044 | membrane organization and biogenesis | 4 | 0.65 | 4.0e-03 |
| GO:0044238 | primary metabolism | 104 | 83.96 | 4.8e-03 |
| GO:0009651 | response to salt stress | 7 | 2.19 | 6.5e-03 |
| GO:0050875 | cellular physiological process | 139 | 119.44 | 7.8e-03 |
| GO:0044237 | cellular metabolism | 111 | 92.12 | 8.2e-03 |
| GO:0009737 | response to abscisic acid stimulus | 7 | 2.34 | 9.3e-03 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0005634 | nucleus | 52 | 24 | 5.7e-08 |
|
list of genes (text format)
Pattern contains 51 genes
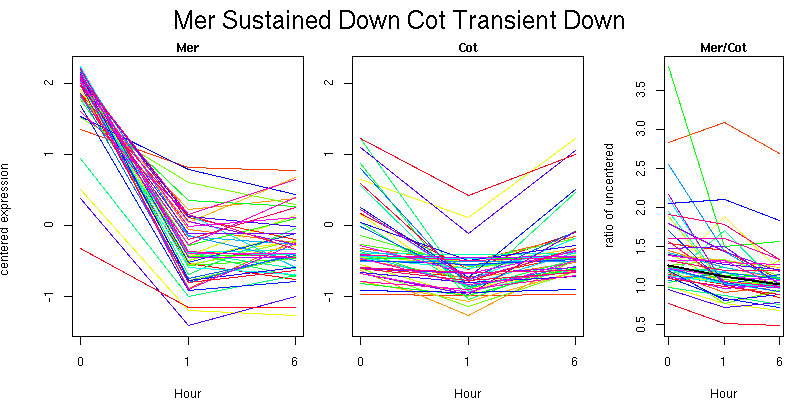
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0030528 | transcription regulator activity | 18 | 4.0 | 2.8e-08 |
| GO:0003700 | transcription factor activity | 16 | 3.7 | 2.7e-07 |
| GO:0003677 | DNA binding | 16 | 4.8 | 9.4e-06 |
| GO:0003676 | nucleic acid binding | 16 | 6.7 | 5.3e-04 |
| GO:0005488 | binding | 25 | 16.4 | 8.9e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0045449 | regulation of transcription | 13 | 3.38 | 1.8e-05 |
| GO:0019219 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 13 | 3.41 | 2.0e-05 |
| GO:0006355 | regulation of transcription, DNA-dependent | 10 | 1.97 | 2.0e-05 |
| GO:0006351 | transcription, DNA-dependent | 10 | 2.03 | 2.6e-05 |
| GO:0031323 | regulation of cellular metabolism | 13 | 3.51 | 2.6e-05 |
| GO:0019222 | regulation of metabolism | 13 | 3.54 | 2.9e-05 |
| GO:0006350 | transcription | 13 | 3.56 | 3.0e-05 |
| GO:0051244 | regulation of cellular physiological process | 13 | 3.84 | 6.8e-05 |
| GO:0050794 | regulation of cellular process | 13 | 3.86 | 7.2e-05 |
| GO:0050791 | regulation of physiological process | 13 | 3.94 | 8.8e-05 |
| GO:0050789 | regulation of biological process | 13 | 4.22 | 1.8e-04 |
| GO:0009733 | response to auxin stimulus | 4 | 0.51 | 1.7e-03 |
| GO:0009719 | response to endogenous stimulus | 7 | 1.76 | 1.7e-03 |
| GO:0006139 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 13 | 5.46 | 2.1e-03 |
| GO:0042221 | response to chemical stimulus | 7 | 2.26 | 6.8e-03 |
| GO:0009725 | response to hormone stimulus | 5 | 1.24 | 7.8e-03 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0005634 | nucleus | 14 | 3.7 | 7.7e-06 |
|
list of genes (text format)
Pattern contains 187 genes
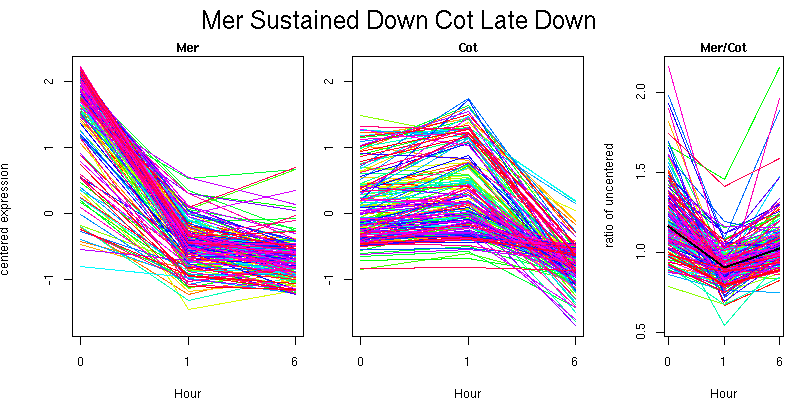
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003824 | catalytic activity | 91 | 57.17 | 5.7e-08 |
| GO:0016903 | oxidoreductase activity, acting on the aldehyde or oxo group of donors | 4 | 0.45 | 1.1e-03 |
| GO:0016491 | oxidoreductase activity | 21 | 10.37 | 1.6e-03 |
| GO:0016787 | hydrolase activity | 30 | 18.90 | 7.0e-03 |
| GO:0016772 | transferase activity, transferring phosphorus-containing groups | 21 | 11.84 | 7.4e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009081 | branched chain family amino acid metabolism | 4 | 0.21 | 5.7e-05 |
| GO:0007582 | physiological process | 108 | 85.77 | 2.7e-04 |
| GO:0008152 | metabolism | 89 | 67.42 | 4.2e-04 |
| GO:0009063 | amino acid catabolism | 4 | 0.36 | 4.4e-04 |
| GO:0009310 | amine catabolism | 4 | 0.36 | 4.4e-04 |
| GO:0044270 | nitrogen compound catabolism | 4 | 0.39 | 6.4e-04 |
| GO:0009605 | response to external stimulus | 8 | 2.17 | 1.5e-03 |
| GO:0042157 | lipoprotein metabolism | 10 | 3.23 | 1.6e-03 |
| GO:0042158 | lipoprotein biosynthesis | 10 | 3.23 | 1.6e-03 |
| GO:0006497 | protein amino acid lipidation | 10 | 3.23 | 1.6e-03 |
| GO:0009628 | response to abiotic stimulus | 23 | 11.82 | 1.6e-03 |
| GO:0019752 | carboxylic acid metabolism | 12 | 4.59 | 2.3e-03 |
| GO:0006082 | organic acid metabolism | 12 | 4.61 | 2.4e-03 |
| GO:0006520 | amino acid metabolism | 8 | 2.42 | 3.0e-03 |
| GO:0006519 | amino acid and derivative metabolism | 10 | 3.56 | 3.1e-03 |
| GO:0006499 | N-terminal protein myristoylation | 9 | 3.13 | 4.4e-03 |
| GO:0018377 | protein myristoylation | 9 | 3.13 | 4.4e-03 |
| GO:0018319 | protein amino acid myristoylation | 9 | 3.13 | 4.4e-03 |
| GO:0043543 | protein amino acid acylation | 9 | 3.14 | 4.4e-03 |
| GO:0009987 | cellular process | 96 | 79.95 | 7.1e-03 |
| GO:0009308 | amine metabolism | 8 | 2.80 | 7.3e-03 |
| GO:0044237 | cellular metabolism | 75 | 59.58 | 8.0e-03 |
| GO:0050875 | cellular physiological process | 93 | 77.25 | 8.2e-03 |
| GO:0043449 | alkene metabolism | 4 | 0.82 | 9.4e-03 |
| GO:0044238 | primary metabolism | 69 | 54.31 | 9.8e-03 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0005622 | intracellular | 89 | 73 | 0.0075 |
|
list of genes (text format)
Pattern contains 356 genes
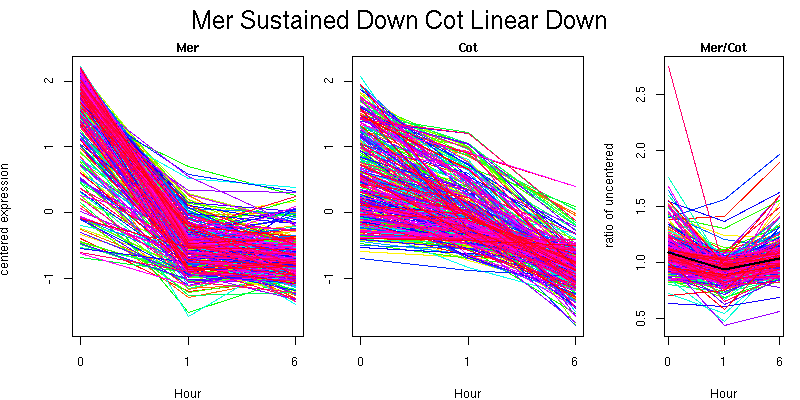
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0008639 | small protein conjugating enzyme activity | 6 | 0.71 | 6.8e-05 |
| GO:0008017 | microtubule binding | 4 | 0.37 | 4.5e-04 |
| GO:0015631 | tubulin binding | 4 | 0.44 | 8.8e-04 |
| GO:0004702 | receptor signaling protein serine/threonine kinase activity | 4 | 0.64 | 3.7e-03 |
| GO:0005057 | receptor signaling protein activity | 4 | 0.66 | 4.1e-03 |
| GO:0004840 | ubiquitin conjugating enzyme activity | 4 | 0.66 | 4.1e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0006914 | autophagy | 6 | 0.30 | 3.5e-07 |
| GO:0043412 | biopolymer modification | 43 | 27.92 | 2.9e-03 |
| GO:0006464 | protein modification | 42 | 27.28 | 3.3e-03 |
| GO:0006499 | N-terminal protein myristoylation | 13 | 5.92 | 6.9e-03 |
| GO:0018377 | protein myristoylation | 13 | 5.92 | 6.9e-03 |
| GO:0018319 | protein amino acid myristoylation | 13 | 5.92 | 6.9e-03 |
| GO:0009639 | response to red or far red light | 5 | 1.20 | 7.0e-03 |
| GO:0043543 | protein amino acid acylation | 13 | 5.94 | 7.1e-03 |
| GO:0042157 | lipoprotein metabolism | 13 | 6.11 | 8.8e-03 |
| GO:0042158 | lipoprotein biosynthesis | 13 | 6.11 | 8.8e-03 |
| GO:0006497 | protein amino acid lipidation | 13 | 6.11 | 8.8e-03 |
| GO:0043283 | biopolymer metabolism | 53 | 38.42 | 9.1e-03 |
|
list of genes (text format)
Pattern contains 32 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 22 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 7 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 44 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009719 | response to endogenous stimulus | 8 | 1.50 | 9.6e-05 |
| GO:0009628 | response to abiotic stimulus | 10 | 2.83 | 3.5e-04 |
| GO:0042221 | response to chemical stimulus | 8 | 1.92 | 5.2e-04 |
| GO:0009725 | response to hormone stimulus | 6 | 1.05 | 5.8e-04 |
| GO:0009733 | response to auxin stimulus | 4 | 0.43 | 9.1e-04 |
| GO:0050896 | response to stimulus | 11 | 4.16 | 2.0e-03 |
| GO:0006950 | response to stress | 7 | 2.31 | 7.2e-03 |
|
list of genes (text format)
Pattern contains 29 genes
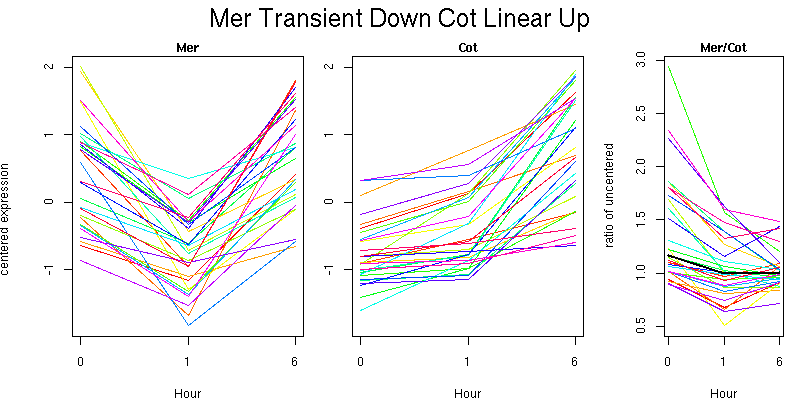
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0031090 | organelle membrane | 4 | 0.8 | 0.008 |
|
list of genes (text format)
Pattern contains 42 genes
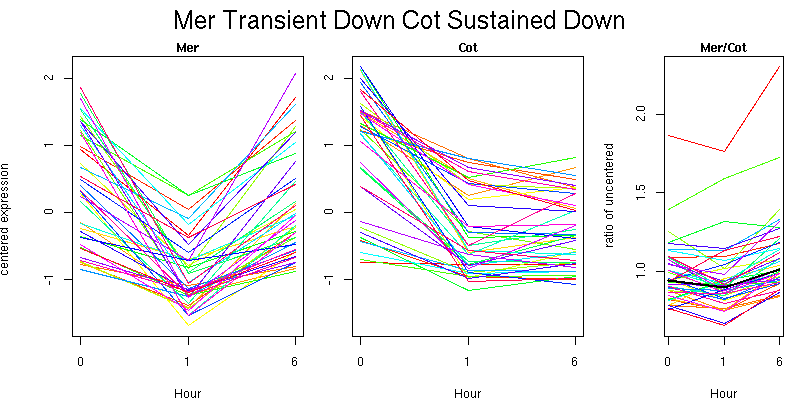
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 44 genes
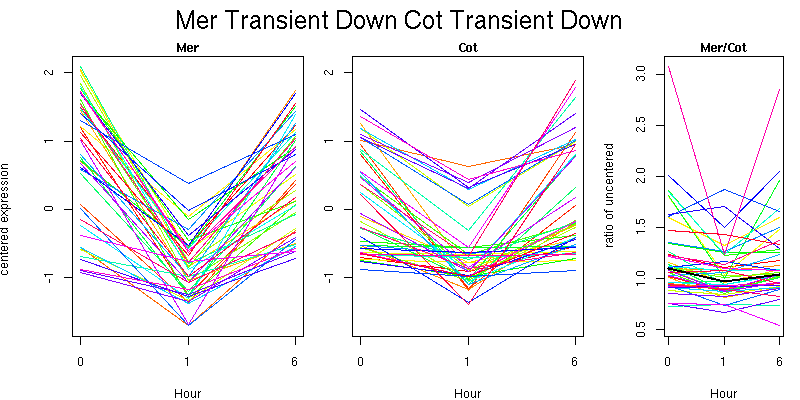
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0008270 | zinc ion binding | 6 | 1.7 | 0.007 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009725 | response to hormone stimulus | 5 | 0.92 | 0.0021 |
| GO:0009719 | response to endogenous stimulus | 5 | 1.30 | 0.0090 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 10 genes
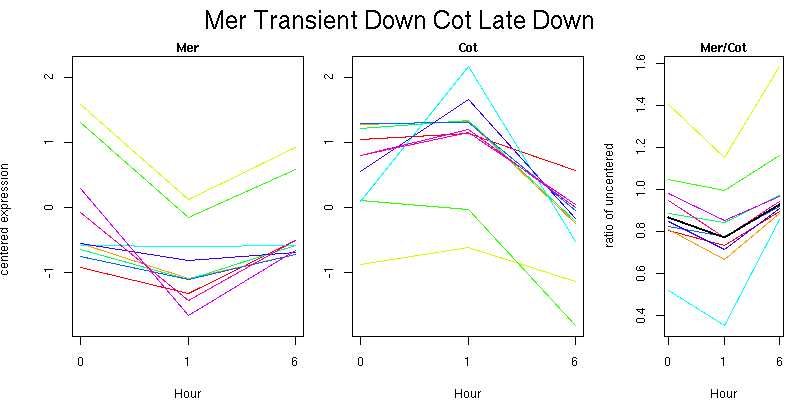
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
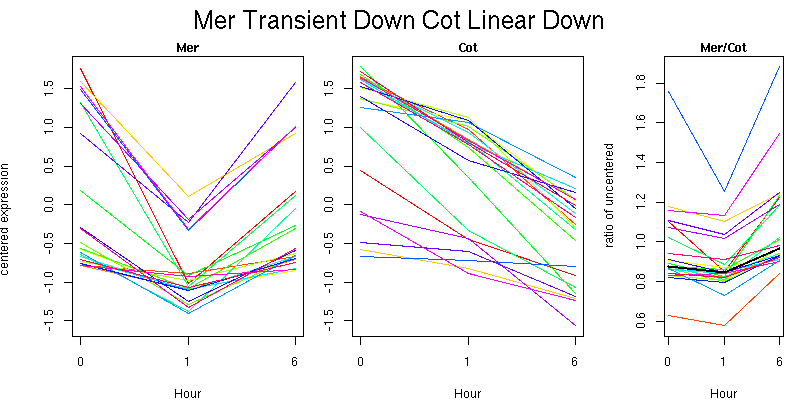
Pattern contains 23 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
list of genes (text format)
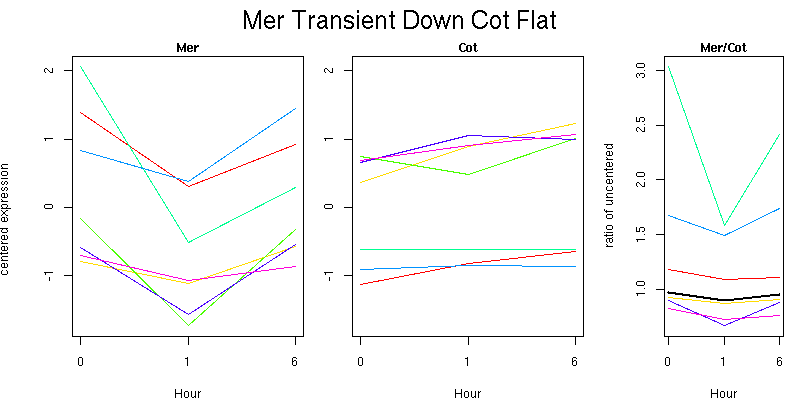
Pattern contains 7 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
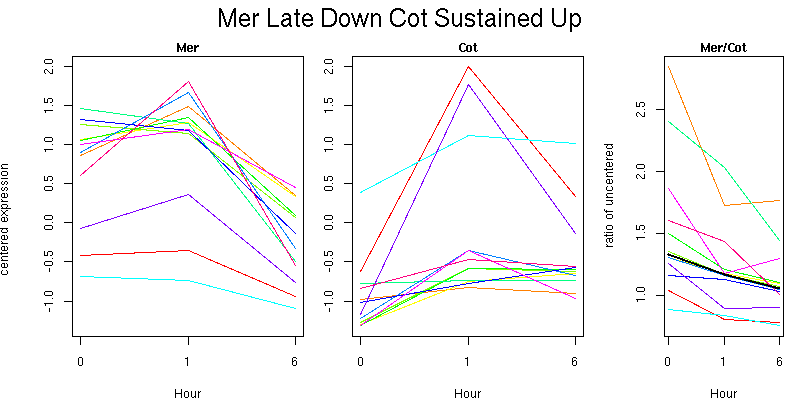
Pattern contains 12 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
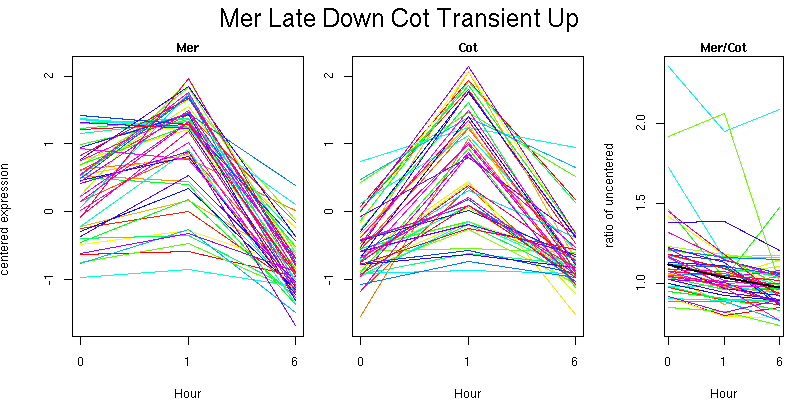
Pattern contains 51 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009737 | response to abscisic acid stimulus | 4 | 0.41 | 0.00073 |
| GO:0009628 | response to abiotic stimulus | 10 | 3.19 | 0.00099 |
| GO:0009719 | response to endogenous stimulus | 7 | 1.69 | 0.00132 |
| GO:0009653 | morphogenesis | 4 | 0.68 | 0.00459 |
| GO:0042221 | response to chemical stimulus | 7 | 2.16 | 0.00530 |
| GO:0050896 | response to stimulus | 11 | 4.69 | 0.00543 |
| GO:0007275 | development | 7 | 2.24 | 0.00636 |
| GO:0009725 | response to hormone stimulus | 5 | 1.19 | 0.00645 |
| GO:0044265 | cellular macromolecule catabolism | 4 | 0.82 | 0.00893 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 5 genes
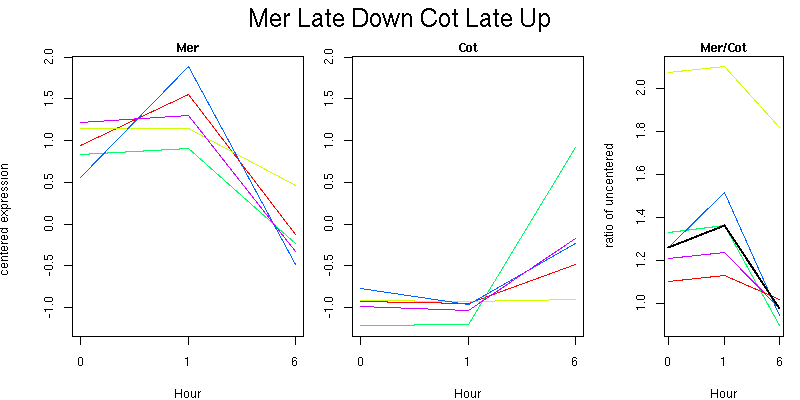
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 9 genes
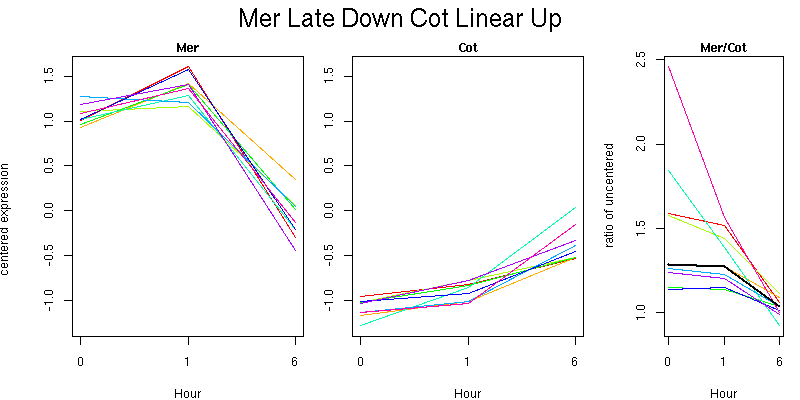
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 12 genes
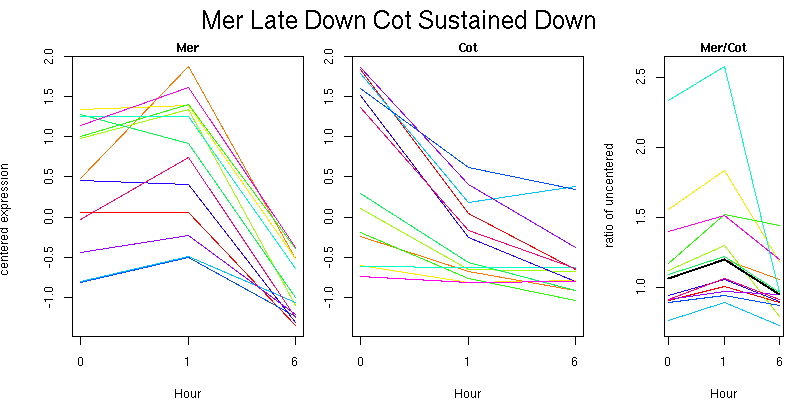
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 2 genes
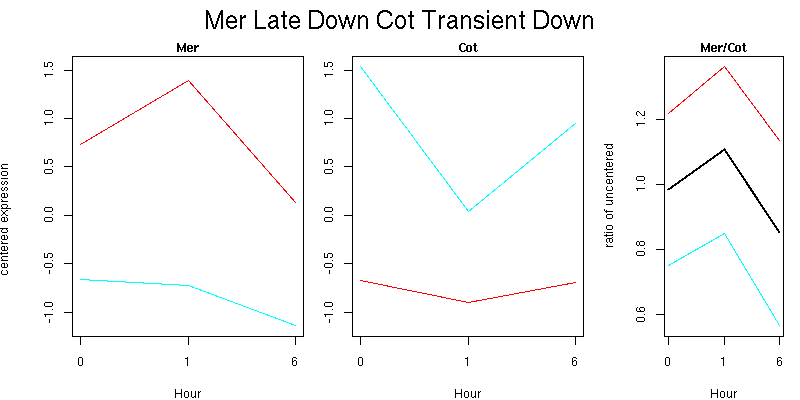
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 134 genes
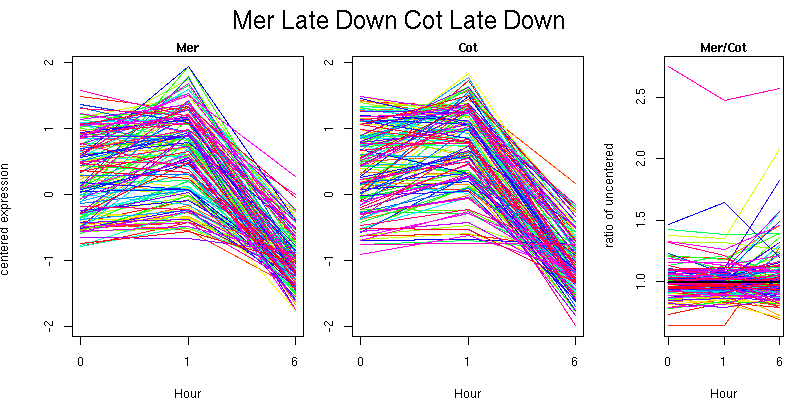
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors | 5 | 0.29 | 1.0e-05 |
| GO:0003824 | catalytic activity | 62 | 40.74 | 4.4e-05 |
| GO:0043492 | ATPase activity, coupled to movement of substances | 6 | 0.88 | 2.6e-04 |
| GO:0042626 | ATPase activity, coupled to transmembrane movement of substances | 6 | 0.88 | 2.6e-04 |
| GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances | 6 | 0.89 | 2.7e-04 |
| GO:0008233 | peptidase activity | 10 | 2.84 | 5.6e-04 |
| GO:0042623 | ATPase activity, coupled | 7 | 1.47 | 7.1e-04 |
| GO:0016491 | oxidoreductase activity | 17 | 7.39 | 1.1e-03 |
| GO:0016887 | ATPase activity | 8 | 2.11 | 1.3e-03 |
| GO:0016787 | hydrolase activity | 25 | 13.47 | 1.6e-03 |
| GO:0017111 | nucleoside-triphosphatase activity | 8 | 2.79 | 7.0e-03 |
| GO:0016462 | pyrophosphatase activity | 8 | 2.92 | 9.0e-03 |
| GO:0016818 | hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides | 8 | 2.92 | 9.2e-03 |
| GO:0016817 | hydrolase activity, acting on acid anhydrides | 8 | 2.93 | 9.3e-03 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0051186 | cofactor metabolism | 10 | 1.380 | 1.3e-06 |
| GO:0009581 | detection of external stimulus | 4 | 0.098 | 2.2e-06 |
| GO:0009737 | response to abscisic acid stimulus | 8 | 1.105 | 1.5e-05 |
| GO:0006818 | hydrogen transport | 5 | 0.340 | 2.2e-05 |
| GO:0015992 | proton transport | 5 | 0.340 | 2.2e-05 |
| GO:0051606 | detection of stimulus | 4 | 0.190 | 3.6e-05 |
| GO:0006732 | coenzyme metabolism | 7 | 0.948 | 4.7e-05 |
| GO:0051188 | cofactor biosynthesis | 6 | 0.739 | 9.8e-05 |
| GO:0009117 | nucleotide metabolism | 6 | 0.791 | 1.4e-04 |
| GO:0050875 | cellular physiological process | 76 | 56.401 | 2.1e-04 |
| GO:0006119 | oxidative phosphorylation | 4 | 0.294 | 2.1e-04 |
| GO:0006752 | group transfer coenzyme metabolism | 4 | 0.327 | 3.2e-04 |
| GO:0006091 | generation of precursor metabolites and energy | 14 | 4.819 | 3.3e-04 |
| GO:0009628 | response to abiotic stimulus | 20 | 8.631 | 3.4e-04 |
| GO:0009725 | response to hormone stimulus | 11 | 3.217 | 3.8e-04 |
| GO:0009987 | cellular process | 77 | 58.369 | 4.0e-04 |
| GO:0007582 | physiological process | 81 | 62.619 | 4.3e-04 |
| GO:0042221 | response to chemical stimulus | 15 | 5.852 | 7.4e-04 |
| GO:0006810 | transport | 24 | 12.116 | 8.2e-04 |
| GO:0051179 | localization | 24 | 12.194 | 9.0e-04 |
| GO:0051234 | establishment of localization | 24 | 12.194 | 9.0e-04 |
| GO:0015672 | monovalent inorganic cation transport | 5 | 0.752 | 9.5e-04 |
| GO:0006508 | proteolysis | 11 | 3.635 | 1.1e-03 |
| GO:0009108 | coenzyme biosynthesis | 4 | 0.451 | 1.1e-03 |
| GO:0050896 | response to stimulus | 24 | 12.691 | 1.6e-03 |
| GO:0019752 | carboxylic acid metabolism | 10 | 3.354 | 2.0e-03 |
| GO:0006082 | organic acid metabolism | 10 | 3.367 | 2.0e-03 |
| GO:0046483 | heterocycle metabolism | 5 | 0.961 | 2.8e-03 |
| GO:0009755 | hormone-mediated signaling | 5 | 1.007 | 3.4e-03 |
| GO:0008152 | metabolism | 64 | 49.222 | 4.1e-03 |
| GO:0009605 | response to external stimulus | 6 | 1.582 | 5.2e-03 |
| GO:0006811 | ion transport | 7 | 2.158 | 6.0e-03 |
| GO:0009719 | response to endogenous stimulus | 11 | 4.564 | 6.0e-03 |
| GO:0044248 | cellular catabolism | 9 | 3.380 | 6.9e-03 |
| GO:0044237 | cellular metabolism | 57 | 43.500 | 7.2e-03 |
| GO:0044249 | cellular biosynthesis | 20 | 11.213 | 7.6e-03 |
| GO:0009056 | catabolism | 9 | 3.492 | 8.4e-03 |
| GO:0015980 | energy derivation by oxidation of organic compounds | 5 | 1.288 | 9.5e-03 |
| GO:0006812 | cation transport | 6 | 1.811 | 9.8e-03 |
| GO:0006092 | main pathways of carbohydrate metabolism | 4 | 0.837 | 1.0e-02 |
|
list of genes (text format)
Pattern contains 111 genes
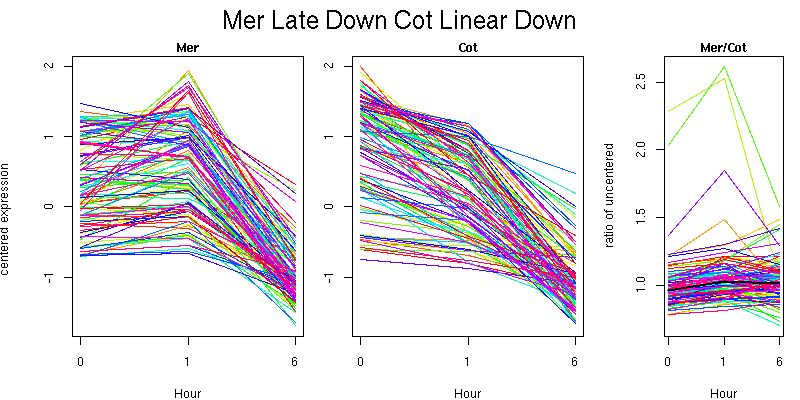
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005525 | GTP binding | 6 | 1.0 | 0.00055 |
| GO:0019001 | guanyl nucleotide binding | 6 | 1.0 | 0.00060 |
| GO:0003824 | catalytic activity | 47 | 34.2 | 0.00548 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 8 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 5 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 36 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0030528 | transcription regulator activity | 8 | 2.5 | 0.0026 |
| GO:0003700 | transcription factor activity | 7 | 2.3 | 0.0061 |
| GO:0003677 | DNA binding | 8 | 3.0 | 0.0072 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0042221 | response to chemical stimulus | 7 | 1.48 | 0.00051 |
| GO:0009725 | response to hormone stimulus | 5 | 0.81 | 0.00115 |
| GO:0009628 | response to abiotic stimulus | 7 | 2.18 | 0.00481 |
| GO:0045449 | regulation of transcription | 7 | 2.21 | 0.00521 |
| GO:0009719 | response to endogenous stimulus | 5 | 1.15 | 0.00526 |
| GO:0019219 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 7 | 2.23 | 0.00545 |
| GO:0031323 | regulation of cellular metabolism | 7 | 2.29 | 0.00631 |
| GO:0019222 | regulation of metabolism | 7 | 2.31 | 0.00664 |
| GO:0006350 | transcription | 7 | 2.32 | 0.00682 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 22 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0050875 | cellular physiological process | 16 | 9.0 | 0.0011 |
| GO:0009987 | cellular process | 16 | 9.3 | 0.0017 |
| GO:0007582 | physiological process | 16 | 10.0 | 0.0043 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0043234 | protein complex | 5 | 1.2 | 0.0064 |
|
list of genes (text format)
Pattern contains 13 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0007047 | cell wall organization and biogenesis | 4 | 0.12 | 4.0e-06 |
| GO:0045229 | external encapsulating structure organization and biogenesis | 4 | 0.12 | 4.0e-06 |
| GO:0016043 | cell organization and biogenesis | 5 | 0.69 | 3.6e-04 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0044425 | membrane part | 4 | 0.56 | 0.0018 |
|
list of genes (text format)
Pattern contains 69 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003723 | RNA binding | 7 | 1.3 | 0.00025 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0046907 | intracellular transport | 6 | 1.29 | 0.0018 |
| GO:0051649 | establishment of cellular localization | 6 | 1.31 | 0.0019 |
| GO:0051641 | cellular localization | 6 | 1.31 | 0.0019 |
| GO:0006886 | intracellular protein transport | 4 | 0.75 | 0.0068 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
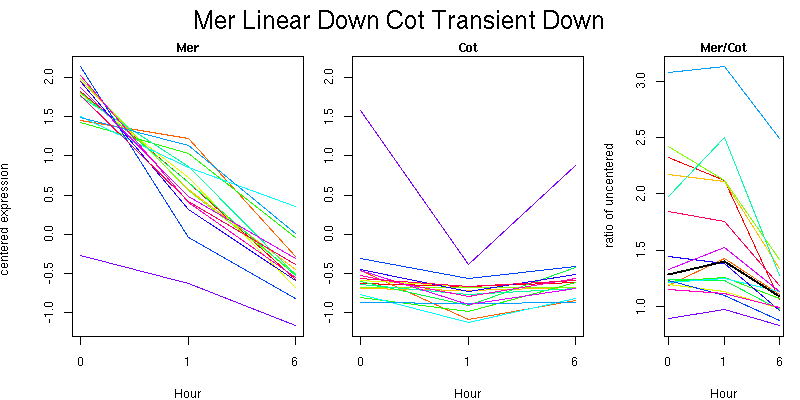
Pattern contains 16 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
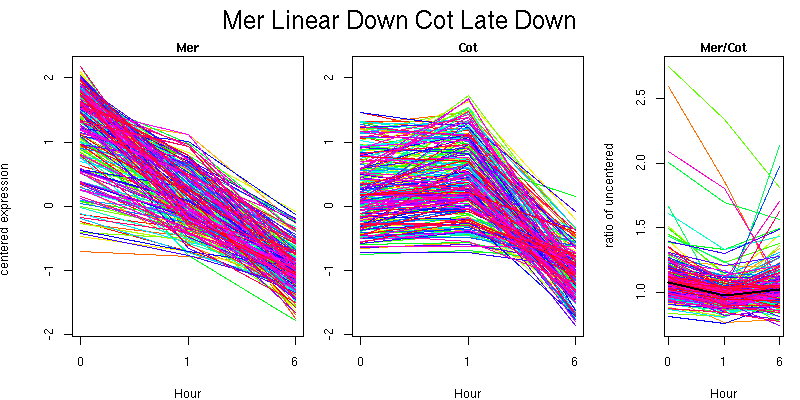
Pattern contains 231 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0005515 | protein binding | 28 | 16 | 0.0031 |
| GO:0003824 | catalytic activity | 92 | 73 | 0.0039 |
|
list of genes (text format)
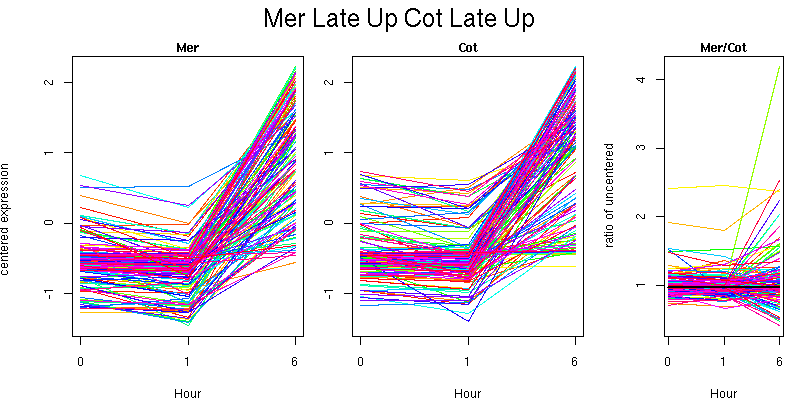
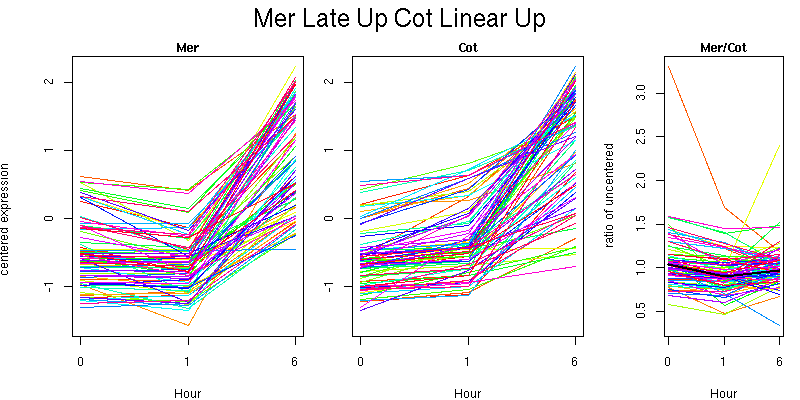
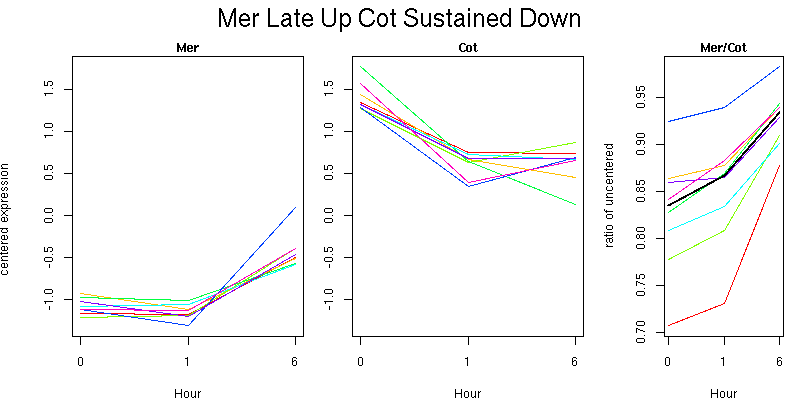
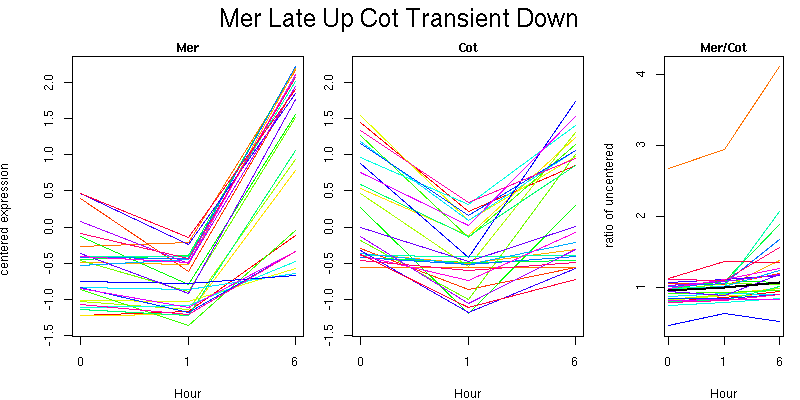
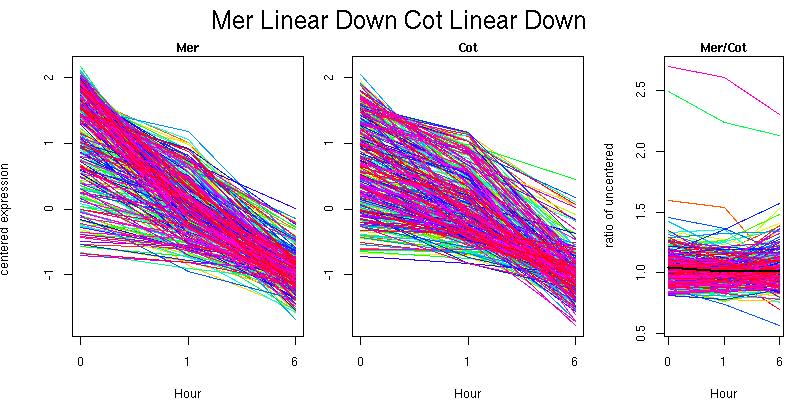
Pattern contains 333 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0008270 | zinc ion binding | 27 | 13.28 | 0.00040 |
| GO:0004840 | ubiquitin conjugating enzyme activity | 4 | 0.63 | 0.00355 |
| GO:0008639 | small protein conjugating enzyme activity | 4 | 0.68 | 0.00465 |
| GO:0016853 | isomerase activity | 8 | 2.77 | 0.00682 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0048518 | positive regulation of biological process | 7 | 1.28 | 0.00029 |
| GO:0008380 | RNA splicing | 4 | 0.50 | 0.00143 |
| GO:0016192 | vesicle-mediated transport | 7 | 1.77 | 0.00205 |
| GO:0006631 | fatty acid metabolism | 7 | 1.85 | 0.00263 |
| GO:0009873 | ethylene mediated signaling pathway | 4 | 0.70 | 0.00526 |
| GO:0048522 | positive regulation of cellular process | 4 | 0.75 | 0.00666 |
| GO:0051242 | positive regulation of cellular physiological process | 4 | 0.75 | 0.00666 |
| GO:0009790 | embryonic development | 10 | 4.00 | 0.00715 |
| GO:0000003 | reproduction | 12 | 5.29 | 0.00724 |
| GO:0048193 | Golgi vesicle transport | 4 | 0.78 | 0.00771 |
|
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0005622 | intracellular | 160 | 134 | 0.0013 |
| GO:0044424 | intracellular part | 154 | 130 | 0.0031 |
| GO:0043227 | membrane-bound organelle | 139 | 118 | 0.0075 |
|
list of genes (text format)
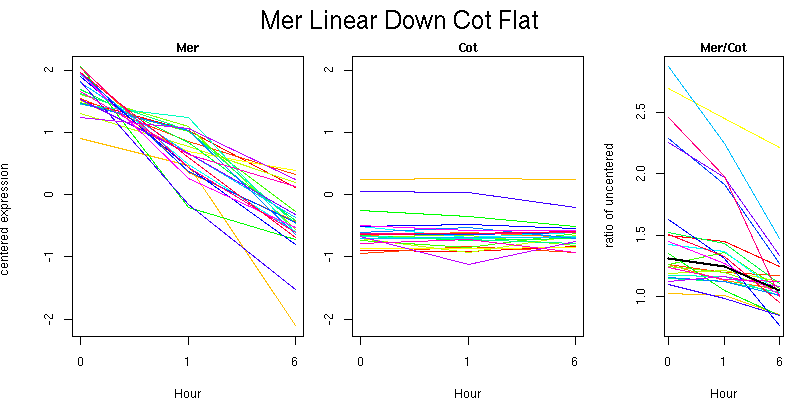
Pattern contains 23 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0009653 | morphogenesis | 4 | 0.32 | 0.00027 |
| GO:0007275 | development | 5 | 1.07 | 0.00347 |
|
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 13 genes
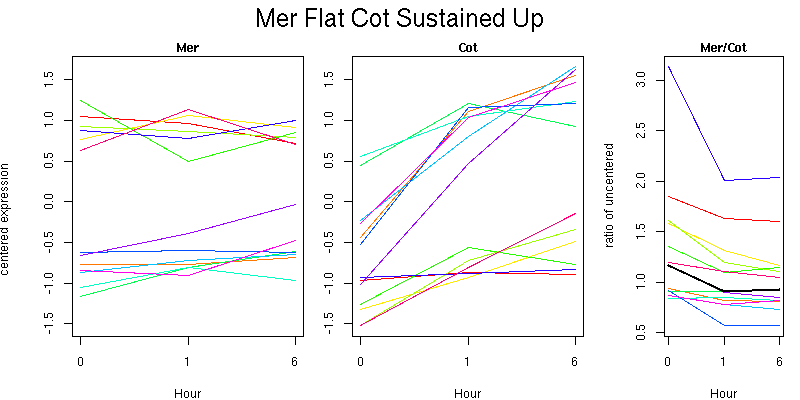
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 16 genes
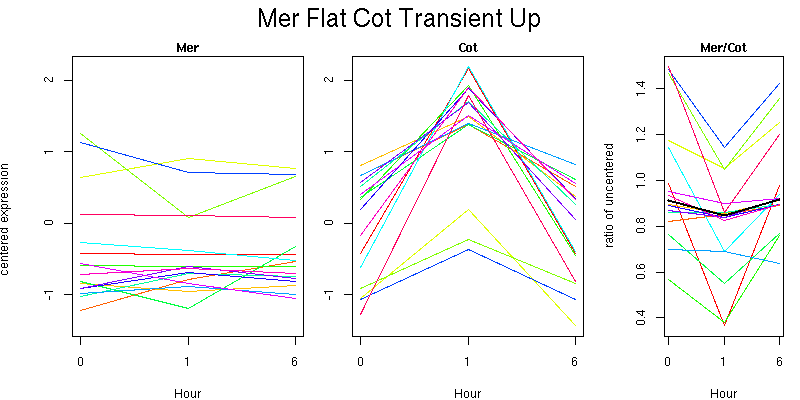
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 16 genes
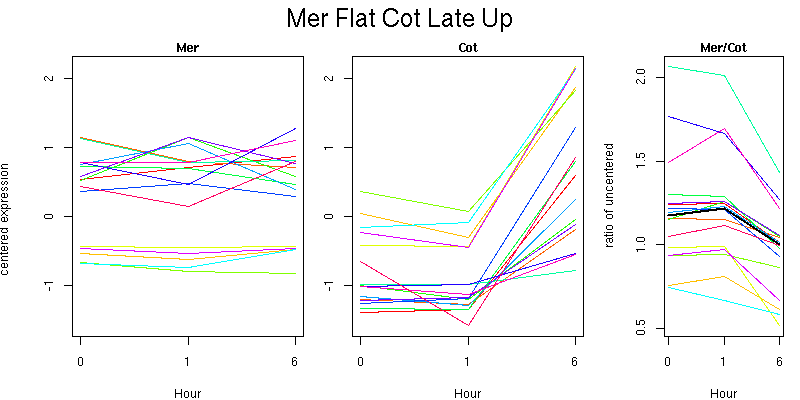
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
Pattern contains 6 genes
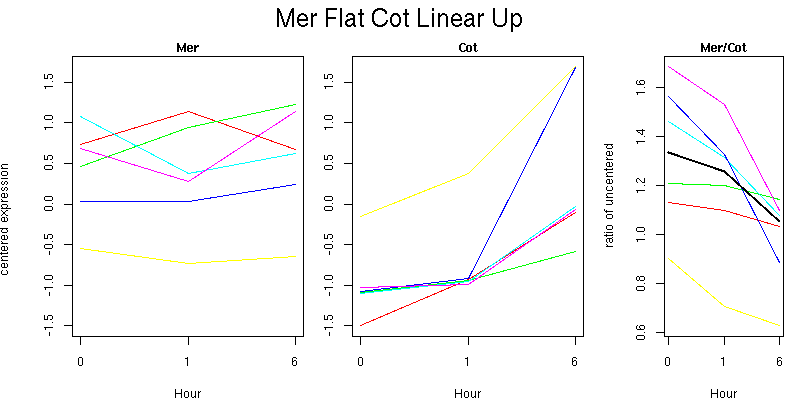
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
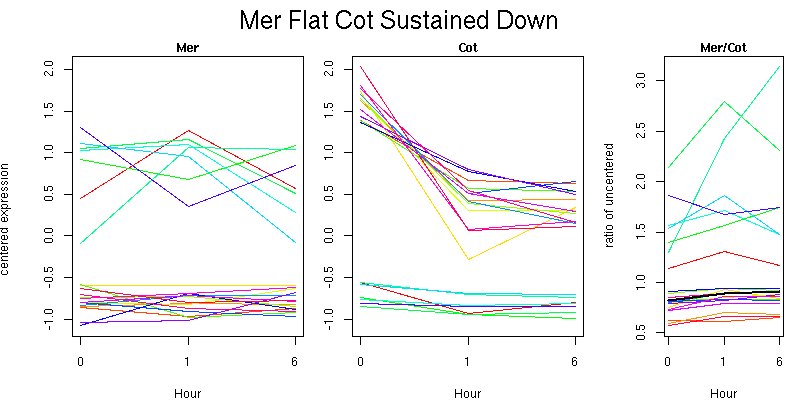
Pattern contains 21 genes
Molecular Function
| |
terms |
counts |
expected.values |
pvals |
| GO:0003700 | transcription factor activity | 6 | 1.5 | 0.0024 |
| GO:0003676 | nucleic acid binding | 8 | 2.7 | 0.0028 |
| GO:0030528 | transcription regulator activity | 6 | 1.6 | 0.0039 |
| GO:0003677 | DNA binding | 6 | 1.9 | 0.0090 |
|
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0019222 | regulation of metabolism | 6 | 1.31 | 0.0012 |
| GO:0050791 | regulation of physiological process | 6 | 1.46 | 0.0021 |
| GO:0050789 | regulation of biological process | 6 | 1.56 | 0.0030 |
| GO:0006355 | regulation of transcription, DNA-dependent | 4 | 0.73 | 0.0051 |
| GO:0006351 | transcription, DNA-dependent | 4 | 0.75 | 0.0057 |
| GO:0045449 | regulation of transcription | 5 | 1.25 | 0.0063 |
| GO:0019219 | regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 5 | 1.26 | 0.0065 |
| GO:0031323 | regulation of cellular metabolism | 5 | 1.30 | 0.0073 |
| GO:0006350 | transcription | 5 | 1.31 | 0.0077 |
|
list of genes (text format)
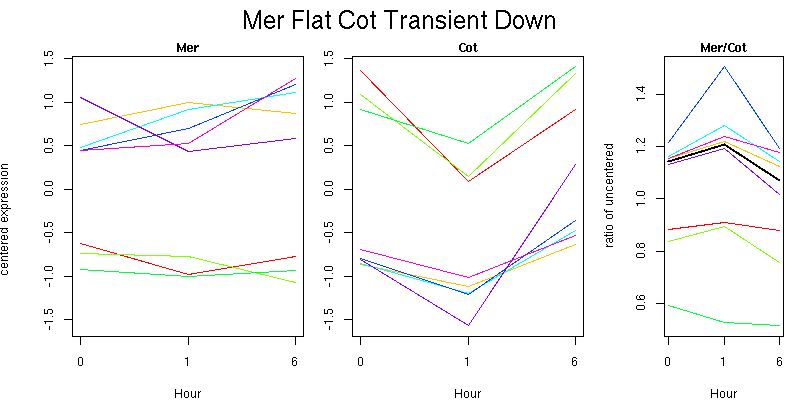
Pattern contains 8 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component - no significant GO terms found
list of genes (text format)
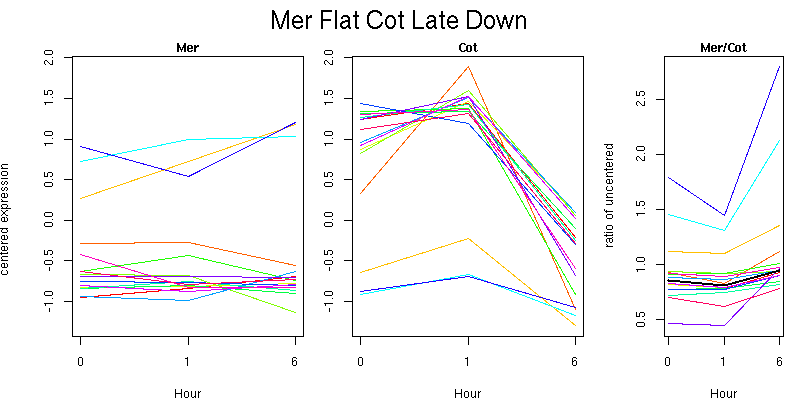
Pattern contains 16 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
list of genes (text format)
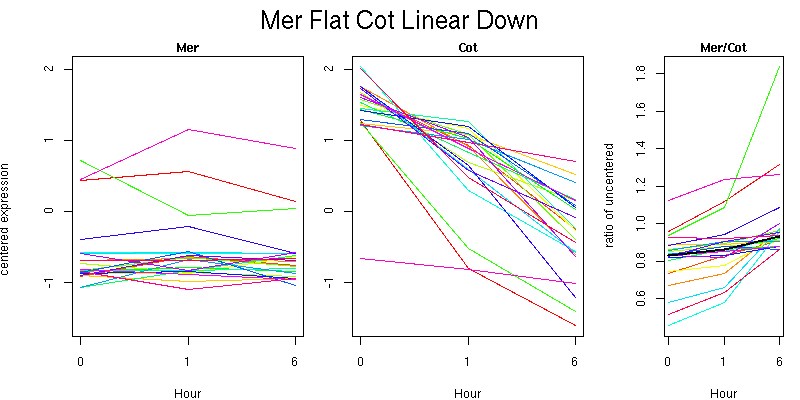
Pattern contains 23 genes
Molecular Function - no significant GO terms found
Biological Pathway - no significant GO terms found
Cellular Component
| |
terms |
counts |
expected.values |
pvals |
| GO:0009536 | plastid | 9 | 3.6 | 0.0047 |
|
list of genes (text format)
Pattern contains 15 genes
Molecular Function - no significant GO terms found
Biological Pathway
| |
terms |
counts |
expected.values |
pvals |
| GO:0019752 | carboxylic acid metabolism | 4 | 0.42 | 0.00066 |
| GO:0006082 | organic acid metabolism | 4 | 0.42 | 0.00067 |
|
list of genes (text format)

















































































