ANNAT1 (ANNEXIN ARABIDOPSIS 1); calcium ion binding / calcium-dependent phospholipid binding
ATTRX2 (Arabidopsis thioredoxin h2, thioredoxin H-type 1); thiol-disulfide exchange intermediate
HYD1 (Hydra 1)
similar to hypothetical protein 13 [Plantago major] (GB:CAJ34821.1)
NA
complex 1 family protein / LVR family protein
GR (GLUTATHIONE REDUCTASE); glutathione-disulfide reductase
OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM C); cysteine synthase
NA
NA
NA
ACR4 (ARABIDOPSIS CRINKLY4); kinase
NA
PTAC17 (PLASTID TRANSCRIPTIONALLY ACTIVE17)
NA
GRF10 (GENERAL REGULATORY FACTOR 10); protein phosphorylated amino acid binding
NA
plastid-lipid associated protein PAP, putative
peroxiredoxin Q, putative
NA
NA
NA
NA
CP29 (chloroplast 29 kDa ribonucleoprotein); RNA binding
ribulose-1,5 bisphosphate carboxylase oxygenase large subunit N-methyltransferase, putative
ATPDR7/PDR7 (PLEIOTROPIC DRUG RESISTANCE 7); ATPase, coupled to transmembrane movement of substances
SET domain-containing protein
cinnamyl-alcohol dehydrogenase, putative
acetylornithine aminotransferase, mitochondrial, putative / acetylornithine transaminase, putative / AOTA, putative / ACOAT, putative
NA
similar to Os01g0894700 [Oryza sativa (japonica cultivar-group)] (GB:NP_001045071.1); similar to P0696G06.13 [Oryza sativa (japonica cultivar-group)] (GB:BAC06256.1); similar to Os09g0515400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063667.1)
NA
peroxiredoxin type 2, putative
peptidase family protein
NA
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G15000.5)
NA
similar to Os06g0320700 [Oryza sativa (japonica cultivar-group)] (GB:NP_001057514.1); similar to hypothetical protein ebA6654 [Azoarcus sp. EbN1] (GB:YP_160826.1); contains InterPro domain Protein of unknown function UPF0061; (InterPro:IPR003846)
AMP binding / acetate-CoA ligase/ catalytic
NA
NA
PDS3 (PHYTOENE DESATURASE)
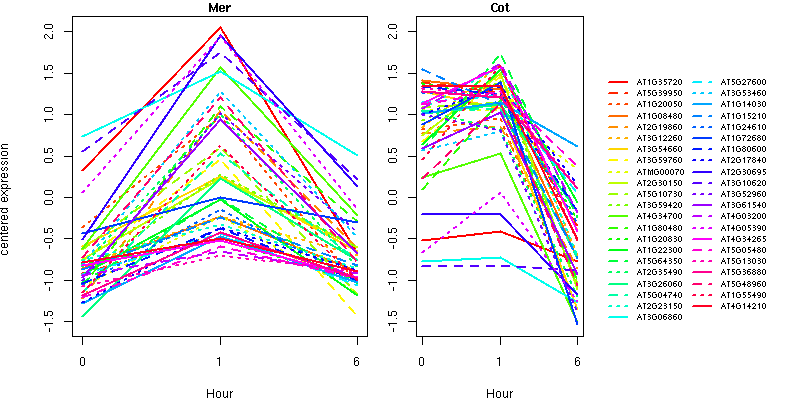
list of genes (text format)