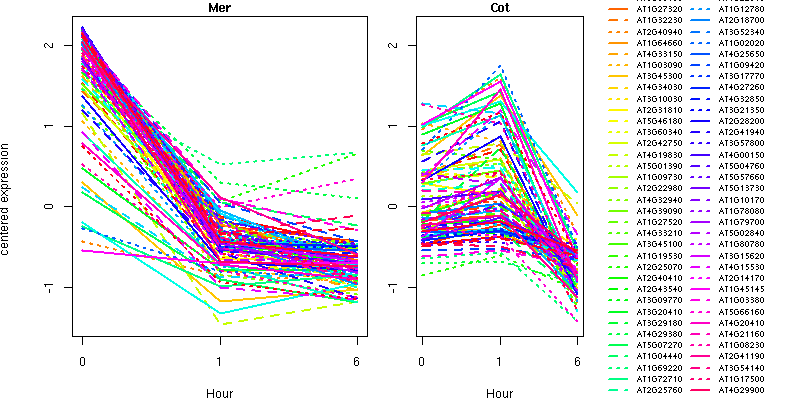
UGE3 (UDP-D-glucose/UDP-D-galactose 4-epimerase 3); UDP-glucose 4-epimerase/ protein dimerization
PHOT2 (NON PHOTOTROPIC HYPOCOTYL 1-LIKE); kinase
ATMRP10 (Arabidopsis thaliana multidrug resistance-associated protein 10)
ATG8B (AUTOPHAGY 8B); microtubule binding
NA
ATMYB33/MYB33 (myb domain protein 33)
AHK3 (ARABIDOPSIS HISTIDINE KINASE 3)
RCD1 (RADICAL-INDUCED CELL DEATH1)
ERS1 (ETHYLENE RESPONSE SENSOR 1); receptor
ATMGL; catalytic/ methionine gamma-lyase
LKR (SACCHAROPINE DEHYDROGENASE)
MCCA (3-methylcrotonyl-CoA carboxylase 1)
IVD (ISOVALERYL-COA-DEHYDROGENASE)
MCCB (3-METHYLCROTONYL-COA CARBOXYLASE); biotin carboxylase
aspartate/glutamate/uridylate kinase family protein
acetolactate synthase small subunit, putative
delta-OAT (ornithine- delta-aminotransferase); ornithine-oxo-acid transaminase
palmitoyl protein thioesterase family protein
NA
immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein
NA
NA
SCPL13; serine carboxypeptidase
GAMMA-VPE (Vacuolar processing enzyme gamma); cysteine-type endopeptidase
NA
glycoside hydrolase family 47 protein
NA
SETH2; transferase, transferring glycosyl groups
NA
NA
NA
NA
NA
CPK9 (CALMODULIN-DOMAIN PROTEIN KINASE 9); calcium- and calmodulin-dependent protein kinase/ kinase
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39430.1); similar to Os12g0577600 [Oryza sativa (japonica cultivar-group)] (GB:NP_001067115.1); similar to Os03g0759000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001051338.1); similar to Protein of unknown function DUF1336 [Medicago truncatula] (GB:ABE78635.1); contains InterPro domain Protein of unknown function DUF1336; (InterPro:IPR009769)
protein kinase family protein / WD-40 repeat family protein
ankyrin repeat family protein
CKL13 (CASEIN KINASE LIKE 13); casein kinase I/ kinase
SIK1 (ERINE/THREONINE KINASE 1); kinase
casein kinase, putative
NA
ankyrin protein kinase, putative
NA
WNK5 (Arabidopsis WNK kinase 5)
CCD1 (CAROTENOID CLEAVAGE DIOXYGENASE 1)
NA
NA
PDS1 (PHYTOENE DESATURATION 1)
|
NA
ACX4 (ACYL-COA OXIDASE 4); oxidoreductase
ETHE1/GLX2-3 (GLYOXALASE 2-3); hydroxyacylglutathione hydrolase
oxidoreductase, 2OG-Fe(II) oxygenase family protein
NA
NA
NA
SPP2 (SPP2); sucrose-phosphatase
nitroreductase family protein
ACD1-LIKE; electron carrier
G6PD4 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE 4); glucose-6-phosphate 1-dehydrogenase
dihydroxyacetone kinase family protein
NA
nPAP (NUCLEAR POLY(A) POLYMERASE); nucleotidyltransferase
NA
nucleic acid binding / transcription factor/ zinc ion binding
NA
NA
scarecrow-like transcription factor 6 (SCL6)
NA
NA
NA
NF-X1 type zinc finger family protein
RAP2.4 (related to AP2 4); DNA binding / transcription factor
NA
DNA binding / transcription factor
CCR4-NOT transcription complex protein, putative
UVR3 (UV REPAIR DEFECTIVE 4)
PPDK (PYRUVATE ORTHOPHOSPHATE DIKINASE)
ALDH6B2 (Aldehyde dehydrogenase 6B2); 3-chloroallyl aldehyde dehydrogenase
ATTRX5 (thioredoxin H-type 5); thiol-disulfide exchange intermediate
AtATG18g (Arabidopsis thaliana homolog of yeast autophagy 18 (ATG18) g)
JR700 (Arabidopsis thaliana receptor homology region transmembrane domain ring H2 motif protein 1); peptidase/ protein binding / zinc ion binding
NA
ZAC (ARF-GAP DOMAIN 12); ARF GTPase activator
similar to amino acid transporter family protein [Arabidopsis thaliana] (TAIR:AT5G41800.1); similar to Os05g0586500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001056462.1); similar to Amino acid/polyamine transporter II [Medicago truncatula] (GB:ABE81500.1); similar to Os01g0857400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001044853.1); contains InterPro domain Amino acid/polyamine transporter II; (InterPro:IPR002422); contains InterPro domain Amino acid transporter, transmembrane; (InterPro:IPR013057)
amino acid transporter family protein
NA
haloacid dehalogenase-like hydrolase family protein
NA
heavy-metal-associated domain-containing protein
exocyst complex subunit Sec15-like family protein
NAP (NAC-LIKE, ACTIVATED BY AP3/PI); transcription factor
MSL3 (MSCS-LIKE 3)
NA
list of genes (text format)