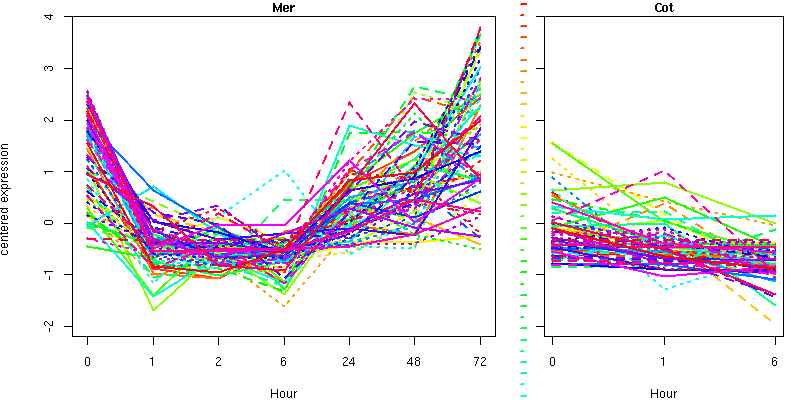
NA
NA
NA
NA
ATMPK6 (MAP KINASE 6); MAP kinase/ kinase
NA
NA
NA
NA
NA
WRKY40 (WRKY DNA-binding protein 40); transcription factor
NA
TRFL1 (TRF-LIKE 1); DNA binding
NA
DNA binding / transcription factor
RCD1 (RADICAL-INDUCED CELL DEATH1)
ATNAC3 (ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 55); transcription factor
NA
AUX1 (AUXIN RESISTANT 1); amino acid permease/ transporter
NA
NA
E2F1; transcription factor
NA
AGO1 (ARGONAUTE 1)
SHY2 (SHORT HYPOCOTYL 2); transcription factor
NA
auxin-responsive family protein
IAA16 (indoleacetic acid-induced protein 16); transcription factor
glutaredoxin family protein
NA
SNRK2-3/SNRK2.3/SRK2I (SNF1-RELATED PROTEIN KINASE 2-3, SNF1-RELATED PROTEIN KINASE 2.3); kinase/ protein kinase
NA
GAI (GA INSENSITIVE); transcription factor
MERI5B (MERISTEM-5); hydrolase, acting on glycosyl bonds
ATMYB33/MYB33 (myb domain protein 33)
ATMPK1 (MITOGEN-ACTIVATED PROTEIN KINASE 1); MAP kinase/ kinase
AHK3 (ARABIDOPSIS HISTIDINE KINASE 3)
NA
ERS1 (ETHYLENE RESPONSE SENSOR 1); receptor
NA
ASN1 (DARK INDUCIBLE 6)
DIN10 (DARK INDUCIBLE 10); hydrolase, hydrolyzing O-glycosyl compounds
NA
UVR3 (UV REPAIR DEFECTIVE 4)
NA
TCH3 (TOUCH 3)
MSL3 (MSCS-LIKE 3)
FRS4 (FAR1-RELATED SEQUENCE 4); zinc ion binding
ERD15 (EARLY RESPONSIVE TO DEHYDRATION 15)
NA
PIF4 (PHYTOCHROME INTERACTING FACTOR 4); DNA binding / transcription factor
NA
CCD1 (CAROTENOID CLEAVAGE DIOXYGENASE 1)
NA
S1 RNA-binding domain-containing protein
SAG21 (SENESCENCE-ASSOCIATED GENE 21)
NA
ALDH6B2 (Aldehyde dehydrogenase 6B2); 3-chloroallyl aldehyde dehydrogenase
NA
ATTRX5 (thioredoxin H-type 5); thiol-disulfide exchange intermediate
ERD5 (EARLY RESPONSIVE TO DEHYDRATION 5, PROLINE OXIDASE); proline dehydrogenase
TRFL6 (TRF-LIKE 6); DNA binding / transcription factor
NA
NA
MATE efflux family protein
ETHE1/GLX2-3 (GLYOXALASE 2-3); hydroxyacylglutathione hydrolase
CAT2 (CATALASE 2); catalase
CAT3 (CATALASE 3); catalase
NA
NA
AtATG18g (Arabidopsis thaliana homolog of yeast autophagy 18 (ATG18) g)
NA
NA
NA
NA
NA
list of genes (text format)