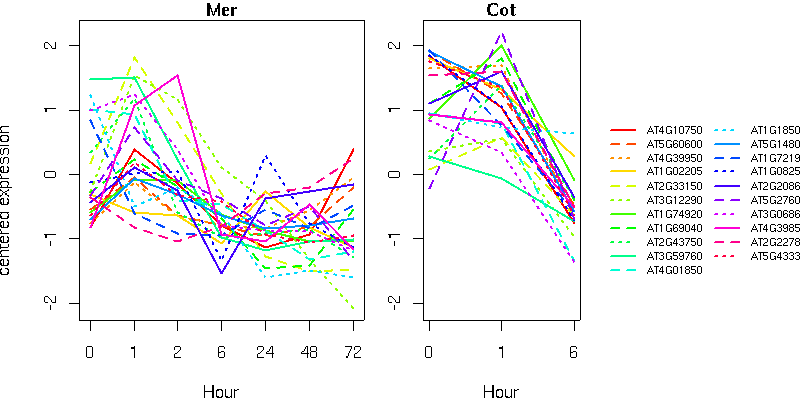
HpcH/HpaI aldolase family protein
GcpE (CHLOROPLAST BIOGENESIS 4); 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase
CYP79B2 (cytochrome P450, family 79, subfamily B, polypeptide 2); oxygen binding
CER1 (ECERIFERUM 1)
NA
tetrahydrofolate dehydrogenase/cyclohydrolase, putative
NA
NA
NA
OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM C); cysteine synthase
MAT2/SAM-2 (S-adenosylmethionine synthetase 2)
NA
P5CR (PYRROLINE-5- CARBOXYLATE (P5C) REDUCTASE)
NA
prephenate dehydratase family protein
LIP1 (Lipoic acid synthase 1); lipoic acid synthase
NA
NA
NA
PMDH1 (PEROXISOMAL NAD-MALATE DEHYDROGENASE 1); malate dehydrogenase
NA
list of genes (text format)