ATIDD4 (ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 4); transcription factor
NA
cytosol aminopeptidase
NA
NA
rac GTPase activating protein, putative
26S proteasome regulatory subunit, putative
NA
NA
CYCA3;2; cyclin-dependent protein kinase
BIK1 (BOTRYTIS-INDUCED KINASE1); kinase
LOS4 (Low expression of osmotically responsive genes 1); ATP-dependent helicase
nuclear RNA-binding protein (RGGA)
NA
NA
APG3 (ALBINO AND PALE GREEN); translation release factor
P5CR (PYRROLINE-5- CARBOXYLATE (P5C) REDUCTASE)
NA
NA
NA
ACR4 (ARABIDOPSIS CRINKLY4); kinase
AP2 domain-containing transcription factor, putative
NA
NA
NA
NA
MYB60 (myb domain protein 60); DNA binding
DNA binding / transcription factor
ATHSFA1D (Arabidopsis thaliana heat shock transcription factor A1D); DNA binding / transcription factor
NA
RNA polymerase II transcription factor/ binding / transcription initiation factor
DNA binding / transcription factor
NA
AP2 domain-containing transcription factor, putative
NA
IAA11 (indoleacetic acid-induced protein 11)
NA
myb family transcription factor
NA
NA
NA
NA
NA
NA
CRU3 (CRUCIFERIN 3)
NA
NA
NA
similar to hypothetical protein 13 [Plantago major] (GB:CAJ34821.1)
endopeptidase Clp
CAT1 (CATALASE 1); catalase
TH1 (THIAMINE REQUIRING 1); phosphomethylpyrimidine kinase/ thiamin-phosphate diphosphorylase
D-CDES (D-CYSTEINE DESULFHYDRASE); 1-aminocyclopropane-1-carboxylate deaminase/ D-cysteine desulfhydrase/ catalytic
NA
NA
NA
NA
electron carrier/ iron ion binding
complex 1 family protein / LVR family protein
ATPQ (ATP SYNTHASE D CHAIN, MITOCHONDRIAL)
NA
GR (GLUTATHIONE REDUCTASE); glutathione-disulfide reductase
OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM C); cysteine synthase
HpcH/HpaI aldolase family protein
NA
methyltransferase
NA
NA
NA
similar to hypothetical protein [Capsicum chinense] (GB:CAJ13713.1)
NA
NA
NA
NA
RAB GTPase activator
monooxygenase
NA
PANB2; 3-methyl-2-oxobutanoate hydroxymethyltransferase
catalytic
EMB1417 (EMBRYO DEFECTIVE 1417)
binding
NA
NA
nucleobase:cation symporter
NA
C2 domain-containing protein
calcium-binding protein, putative
NA
NA
NA
LIP1 (Lipoic acid synthase 1); lipoic acid synthase
ATGPX3 (GLUTATHIONE PEROXIDASE 3); glutathione peroxidase
NA
fructosamine kinase family protein
NA
NA
NA
NA
NA
immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein
NA
ALB4 (ALBINA 4)
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G10000.2); similar to Os08g0558200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001062492.1); similar to unnamed protein product [Ostreococcus tauri] (GB:CAL56846.1); contains InterPro domain Thioredoxin-like fold; (InterPro:IPR012336); contains InterPro domain Thioredoxin fold; (InterPro:IPR012335)
NA
NA
ATNAP7 (Arabidopsis thaliana non-intrinsic ABC protein 7)
HCF109 (HIGH CHLOROPHYLL FLUORESCENT 109); translation release factor
GcpE (CHLOROPLAST BIOGENESIS 4); 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase
nitroreductase family protein
prephenate dehydratase family protein
ATPDR7/PDR7 (PLEIOTROPIC DRUG RESISTANCE 7); ATPase, coupled to transmembrane movement of substances
NA
binding
ATP-dependent protease La (LON) domain-containing protein
NA
methyltransferase
similar to hypothetical protein slr0975 [Synechocystis sp. PCC 6803] (GB:NP_440211.1); similar to Os09g0448900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063318.1)
NA
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35510.1); similar to Os06g0728500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058643.1)
NA
NA
hydrolase
NA
FMN binding
NA
similar to cupin family protein [Arabidopsis thaliana] (TAIR:AT2G18540.1); similar to Os06g0352900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001057589.1); similar to PREDICTED: hypothetical protein, partial [Strongylocentrotus purpuratus] (GB:XP_001185087.1)
similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABA98984.1); similar to Os12g0566100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001067050.1)
NA
calmodulin-related protein, putative
peptidase family protein
NA
NA
NA
APX6 (ASCORBATE PEROXIDASE 6); L-ascorbate peroxidase
NA
LEJ2 (LOSS OF THE TIMING OF ET AND JA BIOSYNTHESIS 2)
NA
NA
pentatricopeptide (PPR) repeat-containing protein
NA
NA
carbohydrate kinase family
similar to Os03g0322600 [Oryza sativa (japonica cultivar-group)] (GB:NP_001049968.1)
NA
NA
AMP binding / acetate-CoA ligase/ catalytic
NA
NA
NA
APO2 (ACCUMULATION OF PHOTOSYSTEM ONE 2)
similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47833.1); similar to Os09g0382500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063065.1)
NA
NA
CYP79B2 (cytochrome P450, family 79, subfamily B, polypeptide 2); oxygen binding
FZL (FZO-LIKE); GTP binding / GTPase/ thiamin-phosphate diphosphorylase
ATOEP16-4; protein translocase
NA
ACD1 (ACCELERATED CELL DEATH 1, PHEOPHORBIDE A OXYGENASE)
NA
NA
cytochrome c oxidase copper chaperone family protein
NA
dynein light chain type 1 family protein
NA
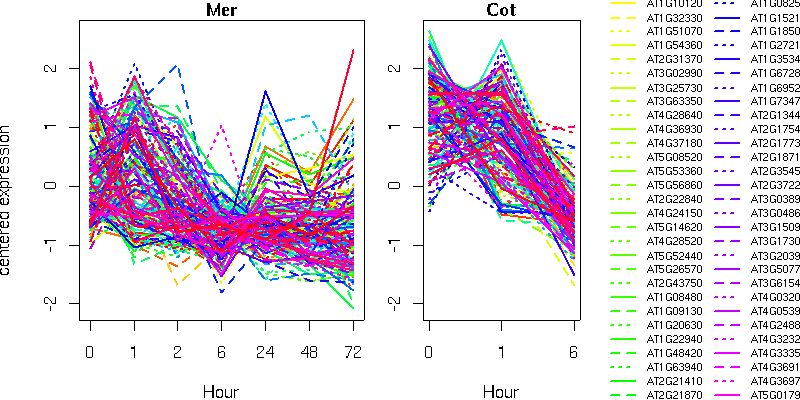
list of genes (text format)